Trans. Nonferrous Met. Soc. China 28(2018) 1838-1846
Response of soil fungal community to long-term chromium contamination
Jin HU1,2, De-long MENG1,2, Xue-duan LIU1,2, Yi-li LIANG1,2, Hua-qun YIN1,2, Hong-wei LIU1,2
1. School of Minerals Processing and Bioengineering, Central South University, Changsha 410083, China;
2. Key Laboratory of Biometallurgy, Ministry of Education, Central South University, Changsha 410083, China
Received 2 December 2017; accepted 29 May 2018
Abstract:
To further study the fungal community in heavy metal contaminated ecosystems, soil samples were collected from an abandoned chromium (Cr) factory, and fungal community was analyzed by Illumina sequencing of Internal Transcribed Spacer (ITS) amplicons. The results showed that Cr contamination changed the composition and structure of soil fungal community, but didn’t change the diversity. Fungus showed various responses to Cr contamination. LEfSe analysis revealed that the biomarker changed a lot in the Cr-contaminated samples in comparison with that in the control samples. The changes in fungal community may be caused by the direct toxic effects on fungi by high concentration of Cr and the significant change in soil properties resulting from Cr contamination. Among all the Cr fractions, organic matter-bound Cr and exchangeable Cr showed significant effects on the fungal community and organic matter also showed a significant effect on soil fungal community.
Key words:
fungal community; alpha-diversity; beta-diversity; Cr contamination; high-throughput sequencing;
1 Introduction
Chromium (Cr) is a chemical element that has been widely applied in many areas such as chemical, factories and foundry. About 85% of the Cr is used in metal alloys [1,2]. The remaining Cr compounds at abandoned industrial sites have high toxic impacts on microorganism, animals and plants [3]. The Cr is an essential micro-nutrient of animals and humans, but high Cr concentration has great harm to animals and humans and causes serious diseases, such as cancer and deformity, or even leads to death. Meanwhile, no positive effects of Cr have been found in plants [4]. CERVANTES et al [4] found that both 0.5 mg/L Cr(VI) solution and 5 mg/L Cr(VI) in soil were harmful to plants. Cr also has great diffusion capacity, because hexavalent chromium, a highly toxic form of Cr, is highly water-soluble and mobile [5]. Soluble Cr can cross the plasma membrane and enter into organisms. The generated free Cr radicals cause direct DNA alterations or other toxic effects [6,7]. CHAI et al [8] have proven that indigenous bacteria in soils could deal with Cr contamination effectively.
Soil microbes are important indicators of soil quality, and heavy metal contamination causes significant changes in the microbial community. Many studies have focused on the responses of bacteria to heavy metal pollution [9-12]. For example, CHAI et al [8] found that the bacteria strains isolated from Cr contaminated soils showed obvious Cr resistance and had effective Cr reduction ability, and our recent study [13] indicated that microbial communities adapt to heavy metal contamination through complex interactions. The fungal community is also an important part of the below-ground ecosystem as the fungi are important organic matter decomposer. Fungal communities cause various responses to heavy metal contamination. For example, VAL et al [14] and LIAO et al [15] found that fungi community composition changed drastically in response to the heavy metals.
However, there are few studies focused on the fungal community in soils facing serious and long-term heavy metal pollution (e.g. the fungal community in soil below the Cr slag). Whereas, understanding the response of fungal community to long-term Cr contamination is of fundamental importance for bioremediation of Cr pollutions.
In the present work, we sampled long-term Cr contaminated soils from an abandoned Cr factory, and investigated soil fungal community using ITS Illumina Miseq sequencing. The aims of this work were to investigate (1) how fungal community responded to the long-term Cr-contamination and (2) which Cr fraction(s) was the major factors affecting fungal community.
2 Experimental
2.1 Site description and soil sampling
Soil samples were gathered in October, 2015. Sampling site is at an abandoned chromium (Cr) salt factory located in Hunan Province, China (E112°58'0", N28°16'23"). The chromate plant has been abandoned for 13 years and no plants are growing in the polluted area. Soil samples (contaminated) were collected (0-20 cm) under the chromate slag heap; meanwhile, control soils were sampled from the adjacent area around the Cr deposit for the same original parent material with contaminated soils (about 500 m far away from the contaminated site and plants can grow). Each type of soil (control and contaminated) was collected three times as the biological replicates. All soil samples were separated into two parts. One part was frozen in liquid nitrogen and stored at -80 °C for further molecular analysis, and the other was brought into the library and was stored at 4 °C for physiochemical property analysis.
Soil physiochemical properties including pH, organic matter and total nitrogen are shown in Table 1, and the Cr contents including exchangeable Cr, Fe/Mn oxides-bound Cr and organic matter-bound Cr were measured according to methods by RAURET et al [16] and MILLER and ZITTEL [17]. Briefly, the exchangeable Cr was extracted using acetate solution, the Fe/Mn oxides-bound Cr was extracted using NH4OH·HCl and HNO3 solution and the organic matter-bound Cr was extracted using H2O2 and NH4OAc solution. The concentration of extracted Cr was measured by the 1,5-diphenylcarbohydrazide spectro- photometric method.
Fungal colony form units (CFU) were determined as described by SIEUWERTS et al [18].
2.2 DNA extraction, PCR and sequencing
DNA was extracted from 10 g soils by DNA extraction method [19]. Then, the quantity and quality of extracted DNA were checked by using a NanoDrop ND-100 spectrophotometer and agarose gel electrophoresis.
PCR amplification was performed on Applied Biosystems 2720 Thermal Cycler (ABI Inc., USA) using ITS primer pair gITS7F (5’-GTG ART CAT CGA RTC TTT G-3’) and ITS4R (5’-TCC TCC GCT TAT TGA TAT GC -3’) together with barcodes and Illumina adapter sequences. Amplification system included 0.5 μL of Taq polymerase (TaKaRa, Japan), 5 μL of 10× PCR buffer, 1.5 μL of dNTP mix, 1.5 μL each primer (10 μmol/L, forward and reverse), 2 μL of DNA template (~20 ng/μL) and 38 μL of deionised H2O. The PCR program was configured as follows: denaturation at 94 °C for 5 min, and 35 cycles of 94 °C for 20 s, 57 °C for 25 s, and 68 °C for 45 s, with a final extension at 68 °C for 10 min. PCR products were purified using E.Z.N.A. TM Gel Extraction Kit (OMEGA Bio-tek Inc., Doraville, GA, USA). Then, the concentration and quality of recovered DNA were evaluated by NanoDrop ND-100 Spectrophotometer (NanoDrop Technologies, Wilmington, USA). For constructing the sequencing library, 200 ng of each purified DNA product was mixed together, and the sequencing was performed on Illumina Miseq platform (Illumina Inc., San Diego, CA, USA).
2.3 Data processing and statistical analysis
Illumina sequencing data were analyzed by an in-house Galaxy pipeline established in ZHOU’s lab (http://zhoulab5.rccc.ou.edu/). Briefly, low quality (QC score <20) sequences were trimmed by Btrim [20] and reads were assigned to samples according to the barcode sequences. Left and right reads were combined by FLASH [21]. OTU assignment was performed using the UPARSE [22] at 97% similarity level. Taxonomic classification was determined with the RDP database at 50% threshold [23]. Raw data sequences were submitted to NCBI SRA database, under the accession number of SAMN07533343, SAMN07533344, SAMN07533345, SAMN07533346, SAMN07533347 and SAMN07533348.
Statistical analyses of fungal community profiles were performed using the STAMP software [24] following the two-sided Welch’s t-test. Alpha-diversity and beta-diversity of fungal community were calculated on the R statistical platform, using the ‘vegan’ package. Alpha-diversity included observed OTU number, Chao1, Shannon diversity index (H), Simpson index of diversity (1/D) and Pielou evenness index (J). Beta-diversity of comparing fungal community structure in different treatments including correspondence analysis (CA), nonmetric multidimensional scaling (NMDs) and analysis of similarity (ANOSIM) were carried out based on Bray-Curtis distance matrix. Canonical correspondence analysis [25] was employed to reveal the relationship between soil properties and fungal community. LEfSe (linear discriminant analysis effect size) for detecting biomarkers was performed on the online Galaxy platform (http://huttenhower.sph.harvard. edu/galaxy/). Student t-test was performed to compare the significant level of difference between two groups using the Minitab software. A p-value of less than 0.05 was considered as significant.
3 Results
3.1 Soil properties
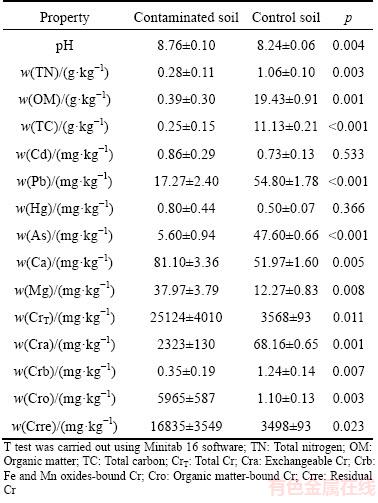
Soil properties such as pH, total nitrogen (TN), organic matter (OM) and different forms of chromate in control and contaminated soils are shown in Table 1. The soils were in alkalinity condition and the contaminated soils had higher pH than the control soils. The main forms of contaminated soils were exchangeable Cr, organic matter-bound Cr and residual Cr, while the Fe/Mn oxides-bound Cr was much less in contaminated soils in comparison with other Cr forms. The Cr content in control soils was high, but the main form was the residual Cr. Total nitrogen (TN) and organic matter (OM) in solids decreased significantly following the Cr contamination.
Table 1 Soil properties of Cr-contaminated and control soils
3.2 Alpha-diversity and abundance of fungal community
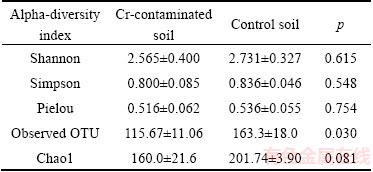
The high-throughput sequencing of ITS rRNA amplicons obtained 110822 clean sequences for 6 samples. To eliminate the effects caused by different sequencing depth, sequencing depth was normalized to 9000 for each sample. Rarefaction curve showed that increasing the sequencing depth would not cause an obvious increase in observed OTU number, which indicated that the sequencing depth is adequate for further analysis (Fig. 1). The rarefaction curve also indicated that control samples had more OTUs than contaminated samples. Alpha-diversity indexes are shown in Table 2. The observed OTU number showed significant (p<0.05) difference between the two groups, while other indexes (Shannon diversity, Simpson diversity, Evenness and Chao1) didn’t.
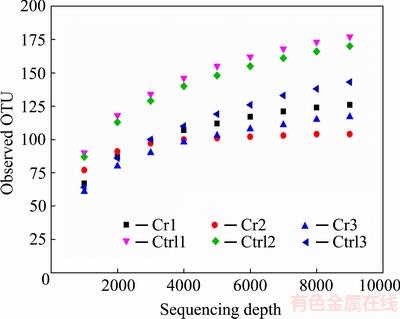
Fig. 1 Observed OTU of Illumina sequencing
Table 2 Fungal community alpha-diversity indexes in Cr- contaminated and control soils
The abundance of the fungal community was determined by colony forming units. The CFU in contaminated-soils ((3.70±1.44)×103 CFU) was 30 times lower than that in control soils ((103.79±15.26)×103 CFU), suggesting that there were much less culturable fungi in contaminated soils.
3.3 Beta-diversity of fungal community
Beta-diversity analyses based on OTU table were carried out by CA, NMDs and ANOSIM analysis. Replicated samples in each treatment grouped well in CA and NMDs plots (Figs. 2(a) and (b)). Dissimilarity analysis (ANOSIM) suggested that the distance between two groups was larger than that within the groups. The difference was not statistically significant (p=0.1), because there were limited replicates (n=3). Heatmap based on active OTUs showed that samples were well clustered, which further revealed the difference in fungal community composition between two groups.
3.4 Phylogenetic composition of fungal community
All sequences could be classified into 5 phyla and 205 genera. The distribution pattern of fungal community at genus level differed obviously between control and contaminated soils (Fig. 3). The genus Candida had the largest abundance in Cr-contaminated samples, while the dominant genus in control samples was the Derxomyces that was rare in contaminated samples.
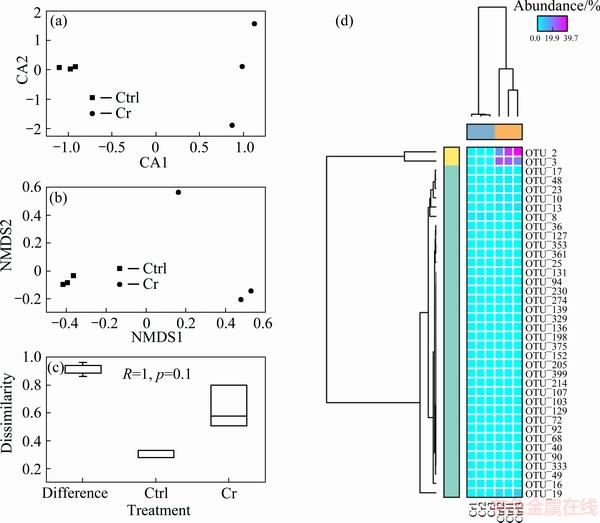
Fig. 2 CA analysis (a), NMDS analysis (b), ANOSIM analysis (c), and heatmap (d) showing fungal community composition difference in Cr-contaminated and control soil samples
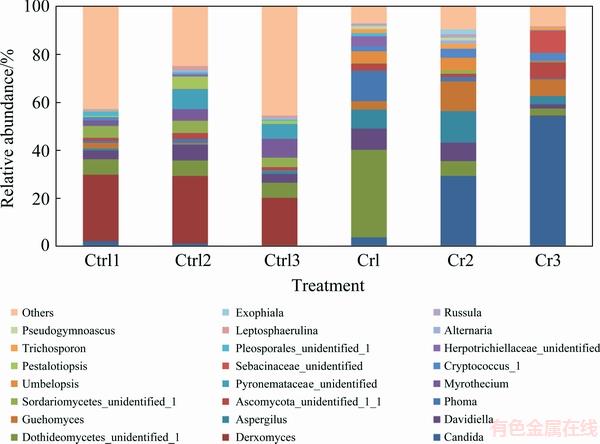
Fig. 3 Relative abundance of fungal community in Cr-contaminated and control soil samples at genus level

Fig. 4 Fungal taxa showing significant differences between Cr-contaminated and control samples at level of phylum (a), classes (b), orders (c), families (d) and genera (e), following Welch’s t-test
Welch’s t-test (Fig. 4) showed that one phylum (Ascomycota), three classes (Sordariomycetes, Orbiliomycetes and Tremellomycetes), six orders (Tremellales, Archaeosporales, Trichosporonales, Teloschistales, Hypocreales and Trichosphaeriales), ten families (Incertae_sedis_12, Archaeosporaceae, Tricho- sporonaceae, Caliciaceae, Vibrisseaceae, Apiosporaceae, Incertae_sedis_3, Parmeliaceae, Elaphomycetaceae and Incertae_sedis_26) and eleven genera (Derxomyces, Trichosporon, Dirinaria, Purpureocillium, Phialo- cephala, Arthrinium, Entoloma, Myrothecium, Nigrospora, Remototrachyna and Elaphomyces) showed significant difference in two treatments. LEFSe analysis (Fig. 5) suggested that members in phylum Ascomycota acted as biomarkers in both control and contaminated soils. For example, Genus Aspergillus in Class Eurotiales Aspergillus and Genus Candida in Order Saccharomycetales were the biomarkers in contaminated samples, whereas Genus Myrothecium, Adisciso and Pestalotiopsis in Order Sordariomycetes were the biomarkers in control samples. In addition, Zygomycota played an important role in Cr-contaminated soils. Trichosporpn, Cryptococcus-1 and Guehomyces from the phylum of Basidiomycota were the biomarkers in contaminated soils and Genus Derxomyces was the biomarker in control soils.
3.5 Soil property and fungal community
CCA (Fig. 6) was carried out to analyze the relationship between soil properties and fungal community. Soil pH, organic matter (OM) and three Cr forms were selected to investigate their effects on the fungal community. The results showed that the exchangeable Cr (p=0.026), Organic matter-bound Cr (p=0.046) and soil organic matter (p=0.013) had significant effects on the fungal community, while the effects of Fe/Mn oxides-bound Cr (p=0.078) and pH (p=0.089) on fungal community were not significant.
4 Discussion
4.1 Effects of Cr slag on soil properties
The long-term Cr pollution caused significant changes in soil physiochemical properties, and particularly, soil organic matters were decreased by about 50 times in polluted soils. KONG et al [26] also found that the heavy metal pollution decreased the content of organic carbon and total nitrogen in soils. Because high level of heavy metals was toxic to plants, a low input of plant and root litter was responsible for the reduction in organic carbon in polluted soils. The observation that no plants could grow in the contaminated region further supported the implication above. In addition, the decomposers (e.g. fungi) in soils mineralized the organic matter and metabolized the organic matter to gas (e.g. CO2) or water-soluble chemicals (e.g. ethanol), and therefore, the original organic matter was consumed [27]. Thus, the organic matter (energy resource) decreased severely in Cr seriously-contaminated soils. The decrease in organic matter further caused a slight decrease in soil pH, because no roots could exude the organic acids to decrease the soils pH in the polluted region, and the more the Cr concentration had, the less the organic acids roots exuded [28].
4.2 Effects of Cr slags on soil fungal community
Serious heavy metal (i.e. Cr slags) caused significant change not only in soil properties but also in fungal communities. Both fungal community richness (observed OTU number) and fungal abundance (CFU) in soils were greatly reduced in response to long-term, serious Cr contamination, which indicated that the serious Cr contamination was toxic to most of fungi. Therefore, Cr sensitive fungi could hardly grow in such condition with a high concentration of Cr. This was further proven by the changes in relative abundance of some fungi. For example, the Derxomyces was the predominant genus in control samples, but could be hardly found in Cr-contaminated samples, because the Derxomyces was very sensitive to heavy metals, and long-term, serious Cr contamination caused extinction of this special genus from soils. Same could be applied to other genera that were sensitive to Cr, such as Dirinaria, Purpureocillium, Phialocephala, Arthrinium and Entoloma. Furthermore, the relative abundance of Trichosporon was obviously increased (p<0.05) in Cr contaminated soils. LAZAROVA et al [29] also found that the Trichosporon cutaneum R57 did not show a significant growth diminution under the Cr(VI) concentration of 10 mmol/L, and suggested that Trichosporon cutaneum R57 had Cr-resistant ability. Besides, GEORGIEVA et al [30] indicated that the Trichosporon cutaneum R57 was resistant to Cr(VI) because the strain had the ability to transform the metal ions into complex polymeric compounds. In addition, BAJGAI et al [31] also found that Trichosporon cutaneum R57 could reduce Cr(VI). Beta-diversity at OUT level, i.e. CA, NMDs, UPGMA and ANOSIM, clearly indicated that the fungal community differed obviously between control and contaminated soils. The results suggested that long-term Cr contamination caused significant changes in fungal abundance (number of fungal species) and fungal community composition in soils.
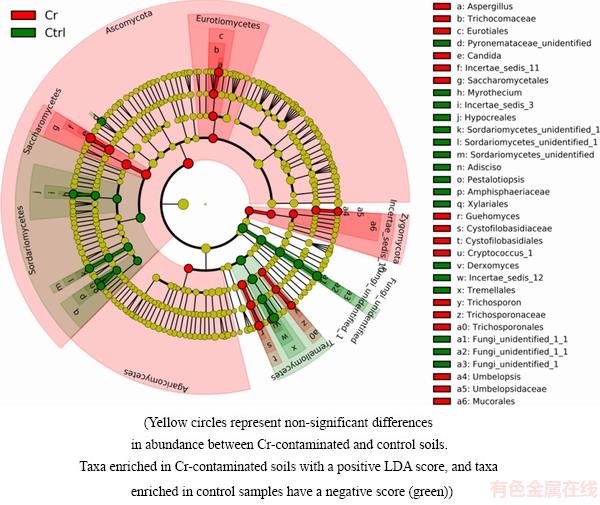
Fig. 5 LEfSe analysis of fungal community in Cr-contaminated and control soils
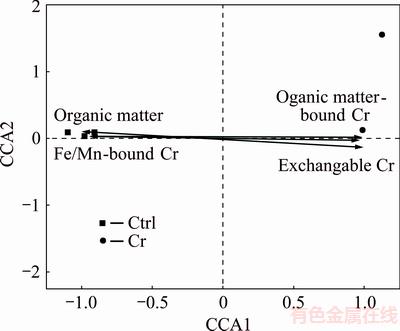
Fig. 6 Canonical correspondence analysis
LEFSE was a reliable method for detecting biomarkers in microbiome [32], and previous studies successfully applied LEFSE in detecting bacterial biomarkers in heavy metal contaminated soils [33]. LEFSE indicated that different taxa acted as the biomarkers in control and contaminated soils. Biomarkers in Cr contaminated soils were supposed to be tolerant to Cr. For example, the genus Candida and the genus Asperillus of phylum Ascomycota were biomarkers in Cr-contaminated samples, and they were both supposed to show obvious heavy metal resistance. For example, the relative abundance of Candida increased obviously in Cr-contaminated samples and many studies suggested that Candida was highly resistant to Cr. BALDI et al [34] suggested that the strain Candida sp. DBVPG 6502 was resistant to Cr. RAMIREZ-RAMIREZ et al [35] found that the strain Candida could grow in medium containing 100 mmol/L Cr, and further proved that the strain had the Cr reduction ability. The Candida was resistant to Cr, because there was NADH-depended chromate reductase in its membrane and protoplast, and the reductase could reduce the toxicity of Cr(VI) [35]. The Aspergillus widely existed in soils [36], and many studies have proven that the Aspergillus was Cr-resistant. For example,  et al [37] indicated that Aspergillus sp. was found in Cr contaminated environments and could reduce Cr(VI) to Cr(III). Similar results that the relative abundance of Aspergillus increased in Cr contaminated samples were obtained in the present work.
et al [37] indicated that Aspergillus sp. was found in Cr contaminated environments and could reduce Cr(VI) to Cr(III). Similar results that the relative abundance of Aspergillus increased in Cr contaminated samples were obtained in the present work.
4.3 Factors constraining soil fungal communities
Because fungi are heterotrophs in soils and take organic matter as the main carbon resource, the soil organic matter is one of the most important factors affecting fungal community [38,39]. In the present work, soil carbon and organic matter were significantly affected by Cr slags. Therefore, soil carbon and organic matter were the main factors constraining fungal community, as indicated by CCA analysis. In addition, GEORGIEVA et al [30] also found that under the high concentration of metal ions, the microorganisms were more difficult to utilize the carbon sources. TEKERLEKOPOULOU et al [40] also suggested that the carbon source was very critical for microbial community structure, as well as the Cr(VI) reduction ability of fungi. Thus, Cr fractions, i.e. organic matter bound Cr, could significantly affect the availability of soil organic matter, and consequently influenced the fungal community. Among all Cr fractions, exchangeable Cr contributed a considerable portion of Cr species and showed the highest effects on fungi community. This is because that the exchangeable Cr is highly water soluble and can easily be absorbed by microorganisms [41,42]. However, the exchangeable Cr is toxic to most of fungi, and therefore, the exchangeable Cr also played important role in regulating fungal community. In contrast to the exchangeable Cr, the Fe/Mn oxides-bound Cr contributed a very low portion of total Cr, and did not show significant effects on fungal community compared with the other Cr forms. It was because that the Fe/Mn oxides bound Cr had very low water solubility and was hard to enter the organism cells, and therefore could not be utilized by microbes [43-46]. Same could also be applied to residual Cr that was water-insoluble and could not be utilized by microorganisms. To summary, organic matter and two Cr fractions, the exchangeable Cr and the organic matter-bound Cr, were the major factors constraining fungal community, whereas, Fe/Mn oxides-bound Cr and residual Cr showed little effects on the fungal community in long-term Cr contaminated soils.
5 Conclusions
1) Cr contamination changed the composition and structure of soil fungal community, but didn’t change the fungal diversity.
2) Cr contamination caused sharp decreases in the number of fungi as well as changes in the fungal community. Biomarkers detected in Cr contaminated soils, for example, the genus Candida and genus Asperillus of phylum Ascomycota were supposed to be highly tolerant to Cr contamination.
3) The changes in the fungal community were caused by the direct toxic effects on fungi caused by high concentration of Cr and the significant changes in soil properties, e.g. change in organic matter, accompanied by Cr contamination. Organic matter was the most important factor that constrained fungal community and among all the Cr fractions organic matter bound Cr and exchangeable Cr showed significant effects on the fungal community.
References
[1] MORRISON R D, MURPHY B L. Environmental forensics: Contaminant specific guide [M]: New York: Elsevier, 2010: 313-337.
[2] CHEN Bian-fang, HUANG Sheng, LIU Biao, GE Qi, XIE Shu-shan, WANG Ming-yu, WANG Xue-wen. Thermodynamic analysis for separation of vanadium and chromium in V(IV)-Cr(III)-H2O system [J]. Transactions of Nonferrous Metals Society of China, 2018, 28: 567-573.
[3] ZHU Feng, HOU Jing-tao, XUE Sheng-guo, WU Chuan, WANG Qiong-li, HARTLEY W. Vermicompost and gypsum amendments improve aggregate formation in bauxite residue [J]. Land Degradation and Development, 2017, 28(7): 2109-2120.
[4] CERVANTES C, CAMPOS-GARCIA J, DEVARS S, GUTIERREZ- CORONA F, LOZA-TAVERA H, TORRES-GUZMAN J C, MORENO-SANCHEZ R. Interactions of chromium with microorganisms and plants [J]. FEMS Microbiology Reviews, 2001, 25: 335-347.
[5] MEGHARAJ M, AVUDAINAYAGAM S, NAIDU R. Toxicity of hexavalent chromium and its reduction by bacteria isolated from soil contaminated with tannery waste [J]. Current Microbiology, 2003, 47: 51-54.
[6] ARSLAN P, BELTRAME M, TOMASI A. Intracellular chromium reduction [J]. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1987, 931: 10-15.
[7] KADIISKA M B, XIANG Qun-Hui, MASON R P. In vivo free radical generation by chromium (VI): An electron spin resonance spin-trapping investigation [J]. Chemical Research in Toxicology, 1994, 7: 800-805.
[8] CHAI Li-yuan, HUANG Shun-hong, YANG Zhi-hui, PENG Bing, HUANG Yan, CHEN Yue-hui. Cr(VI) remediation by indigenous bacteria in soils contaminated by chromium-containing slag [J]. Journal of Hazardous Materials, 2009, 167: 516-522.
[9] SHI W, BECKER J, BISCHOFF M, TURCO R F, KONOPKA A E. Association of microbial community composition and activity with lead, chromium, and hydrocarbon contamination [J]. Applied and Environmental Microbiology, 2002, 68: 3859-3866.
[10] ZHU Feng, CHENG Qing-yu, XUE Sheng-guo, LI Chu-xuan, HARTLEY W, WU Chuan, TIAN Tao. Influence of natural regeneration on fractal features of residue microaggregates in bauxite residue disposal areas [J]. Land Degradation and Development, 2018, 29(1): 138-149.
[11] WANG Jun, YE Sheng, XUE Sheng-guo, HARTLEY W, WU Hao, SHI Li-zheng. The physiological response of Mirabilis jalapa Linn. to lead stress and accumulation [J]. International Biodeterioration and Biodegration, 2018,128: 11-14.
[12] GOLEBIEWSKI M, DEJA-SIKORA E, CICHOSZ M, TRETYN A, WROBEL B. 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils [J]. Microbial Ecology, 2014, 67: 635-647.
[13] LI Xiao-qi, MENG De-long, LI Juan, YIN Hua-qun, LIU Hong-wei, LIU Xue-duan, CHENG Cheng, XIAO Yun-hua, LIU Zheng-hua, YAN Ming-li. Response of soil microbial communities and microbial interactions to long-term heavy metal contamination [J]. Environmental Pollution, 2017, 231(Pt 1): 908-917.
[14] VAL C D, BAREA J M, AZCONAGUILAR C. Diversity of arbuscular mycorrhizal fungus populations in heavy-metal- contaminated soils [J]. Applied and Environmental Microbiology, 1999, 65: 718-723.
[15] NORDGREN A,  B. Microfungi and microbial activity along a heavy metal gradient [J]. Applied and Environmental Microbidogy, 1983, 45: 1829-1837.
B. Microfungi and microbial activity along a heavy metal gradient [J]. Applied and Environmental Microbidogy, 1983, 45: 1829-1837.
[16] RAURET G, LOPEZ-SANCHEZ J F, SAHUQUILLO A, RUBIO R, DAVIDSON C, URE A, QUEVAUVILLER P. Improvement of the BCR three step sequential extraction procedure prior to the certification of new sediment and soil reference materials [J]. Journal of Environmental Monitoring, 1999, 1: 57-61.
[17] MILLER F J, ZITTEL H E. Spectrophotometric Determination of Technetium with 1,5-Diphenylcarbohydrazide [J]. Analytical Chemistry, 1963, 35: 299-301.
[18] SIEUWERTS S, de BOK F A M, MOLS E, de VOS W M, van HYLCKAMA VLIEG J. A simple and fast method for determining colony forming units [J]. Letters in Applied Microbiology, 2010, 47: 275-278.
[19] GABOR E, VRIES E J, JANSSEN D B. Efficient recovery of environmental DNA for expression cloning by indirect extraction methods [J]. FEMS Microbiology Ecology, 2003, 44: 153-163.
[20] KONG Yong. Btrim: A fast, lightweight adapter and quality trimming program for next-generation sequencing technologies [J]. Genomics, 2014, 98: 152-153.
[21] MAGOC T, SALZBERG S L. FLASH: fast length adjustment of short reads to improve genome assemblies [J]. Bioinformatics, 2011, 27: 2957-2963.
[22] EDGAR R. UPARSE: highly accurate OTU sequences from microbial amplicon reads [J]. Nature Methods, 2013, 10: 996-998.
[23] COLE J R, WANG Qiong, CARDENAS E, FISH J, CHAI Ben-li, FARRIS R J, KULAM-SYED-MOHIDEEN A S, McGARRELL D M, MARSH T, GARRITY G M. The Ribosomal database project: Improved alignments and new tools for rRNA analysis [J]. Nucleic Acids Research, 2009, 37: D141-D145.
[24] PARKS D H, TYSON G W, HUGENHOLTZ P, BEIKO R G. STAMP: statistical analysis of taxonomic and functional profiles [J]. Bioinformatics, 2014, 30: 3123-3124.
[25] MCCARTHY A J, WILLIAMS S T. Actinomycetes as agents of biodegradation in the environment—A review [J]. Gene, 1992, 115: 189-192.
[26] KONG Xiang-feng, JIANG Xing-xing, XUE Sheng-guo, HUANG Ling, HARTLEY W, CHUAN Wu, LI Xiao-fei. Migration and distribution of salinity in bauxite residue during water leaching [J]. Transactions of Nonferrous Metals Society of China, 2018, 28: 534-541.
[27] BUSCOT F, VARMA A. Microorganisms in soils: Roles in genesis and functions[M]. Springer, 2005.
[28] KONG Xiang-feng, TIAN Tao, XUE Sheng-guo, HARTLEY W, HUANG Long-bin, WU Chuan, LI Chu-xuan. Development of alkaline electrochemical characteristics demonstrates soil formation in bauxite residue undergoing natural rehabilitation [J]. Land Degradation and Development, 2018, 29(1): 58-67.
[29] LAZAROVA N, KRUMOVA E, STEFANOVA T, GEORGIEVA N, ANGELOVA M. The oxidative stress response of the filamentous yeast Trichosporon cutaneum R57 to copper, cadmium and chromium exposure [J]. Biotechnology and Biotechnological Equipment, 2014, 28: 855-862.
[30] GEORGIEVA N, PESHEV D, RANGELOVA N, LAZAROVA N. Effect of hexavalent chromium on growth of Trichosporon cutaneum R57 [J]. J Univ Chem Technol Metall, 2011, 46: 293-298.
[31] BAJGAI R C, GEORGIEVA N, LAZAROVA N. Bioremediation of chromium ions with filamentous yeast Trichosporon cutaneum R57 [J]. Journal of Biology and Earth Sciences, 2012, 2: 70-75.
[32] SEGATA N, IZARD J, WALDRON L, GEVERS D, MIROPOLSKY L, GARRETT W S, HUTTENHOWER C. Metagenomic biomarker discovery and explanation [J]. Genome Biology, 2011, 12: R60.
[33] XIAO En-zong, KRUMINS V, XIAO Tang-fu, DONG Yi-ran, TANG Song, NING Zeng-ping, HUANG Zheng-yu, SUN Wei-min. Depth-resolved microbial community analyses in two contrasting soil cores contaminated by antimony and arsenic [J]. Environmental Pollution, 2017, 221: 244-255.
[34] BALDI F, VAUGHAN A M, OLSON G J. Chromium(VI)-resistant yeast isolated from a sewage treatment plant receiving tannery wastes [J]. Applied and Environmental Microbiology, 1990, 56: 913-918.
[35] 
 G J. Cr(VI) reduction in a chromate-resistant strain of Candida maltosa isolated from the leather industry [J]. Antonie van Leeuwenhoek, 2004, 85: 63-68.
G J. Cr(VI) reduction in a chromate-resistant strain of Candida maltosa isolated from the leather industry [J]. Antonie van Leeuwenhoek, 2004, 85: 63-68.
[36] LENART-BORO A, BORO P. The effect of industrial heavy metal pollution on microbial abundance and diversity in soils—A review [M]. Intech Europe, 2014.
[37]  M, RASPOR P, SIPICZKI M, PESTI M. Hexavalent chromium uptake by sensitive and tolerant mutants of Schizosaccharomyces pombe [J]. FEMS Microbiology Letters, 1999, 178: 109-115.
M, RASPOR P, SIPICZKI M, PESTI M. Hexavalent chromium uptake by sensitive and tolerant mutants of Schizosaccharomyces pombe [J]. FEMS Microbiology Letters, 1999, 178: 109-115.
[38] LEHMANN J, KLEBER M. The contentious nature of soil organic matter [J]. Nature, 2015, 528: 60-68.
[39] XUE Sheng-guo, LI Meng, JIANG Jun, MILLAR G J, LI Chu-xuan, KONG Xiang-feng. Phosphogypsum stabilization of bauxite residue: conversion of its alkaline characteristics [J]. Journal of Environmental Sciences, 2018: DOI: 10.1016/j.jes.2018.05.016.
[40] TEKERLEKOPOULOU A G, TSIAMIS G, DERMOU E, SIOZIOS S, BOURTZIS K, VAYENAS D V. The effect of carbon source on microbial community structure and Cr(VI) reduction rate [J]. Biotechnology and Bioengneering, 2010, 107: 478-87.
[41] JAMES B R, BARTLETT R J. Behavior of chromium in soils: VII. Adsorption and reduction of hexavalent forms [J]. Journal of Environmental Quality, 1983, 12: 177-181.
[42] MIN Xiao-bo, WANG Yun-yan, CHAI Li-yuan, YANG Zhi-hui, LIAO Ying-ping. High-resolution analyses reveal structural diversity patterns of microbial communities in chromite ore processing residue (COPR) contaminated soils [J]. Chemosphere, 2017, 183: 266-276.
[43] COHEN M D, KARGACIN B, KLEIN C B, COSTA M. Mechanisms of chromium carcinogenicity and toxicity [J]. Critical Reviews in Toxicology, 1993, 23: 255-281.
[44] CHECKAI R T, COREY R B, HELMKE P A. Effects of ionic and complexed metal concentrations on plant uptake of cadmium and micronutrient metals from solution [J]. Plant and Soil, 1987, 99: 335-345.
[45] ZHU Feng, LIAO Jia-xin, XUE Sheng-guo, HARTLEY W, ZOU Qi, WU Hao. Evaluation of aggregate microstructures following natural regeneration in bauxite residue as characterized by synchrotron-based X-ray micro-computed tomography [J]. Science of the Total Environment, 2016, 573: 155-163.
[46] GILLER K E, WITTER E, MCGRATH S P. Toxicity of heavy metals to microorganisms and microbial processes in agricultural soils: A review [J]. Soil Biology and Biochemistry, 1998, 30: 1389-1414.
长期铬污染对土壤真菌群落的影响
胡 瑾1,2,孟德龙1,2,刘学端1,2,梁伊丽1,2,尹华群1,2,刘宏伟1,2
1. 中南大学 资源加工与生物工程学院,长沙 410083;
2. 中南大学 生物冶金教育部重点实验室,长沙 410083
摘 要:为进一步研究重金属污染生态系统中真菌群落的变化,以废弃铬盐厂污染的土壤为研究对象,通过二代测序ITS扩增子技术,研究真菌微生物群落。结果显示,铬污染虽然改变了真菌群落的组成和结构,但是对其多样性并没有显著的影响。不同种类的真菌对铬污染的响应不同。LEfSe分析结果显示,铬污染环境会导致真菌种类的变化,这是由于高浓度的铬污染能够对真菌细胞直接产生毒性,或者通过改变土壤性质,使真菌细胞对土壤中碳、氮等能源物质的利用能力下降。在所有铬的形态中,有机结合态和可交换态的铬对真菌群落的影响最显著;而土壤性质中,有机质对真菌群落的影响最为显著。
关键词:真菌群落;α多样性;β多样性;铬污染;高通量测序
(Edited by Bing YANG)
Foundation item: Project (51504298) supported by the National Natural Science Foundation of China; Project (2016JJ3146) supported by the Natural Science Foundation of Hunan Province, China; Project (1053320171098) supported by the Fundamental Research Funds for the Central Universities of China; Project supported by the Postdoctoral Research Funding Plan in Central South University, China
Corresponding author: Hong-wei LIU; Tel: +86-731-88830546; E-mail: hongweiliu@csu.edu.cn
DOI: 10.1016/S1003-6326(18)64828-9
Abstract: To further study the fungal community in heavy metal contaminated ecosystems, soil samples were collected from an abandoned chromium (Cr) factory, and fungal community was analyzed by Illumina sequencing of Internal Transcribed Spacer (ITS) amplicons. The results showed that Cr contamination changed the composition and structure of soil fungal community, but didn’t change the diversity. Fungus showed various responses to Cr contamination. LEfSe analysis revealed that the biomarker changed a lot in the Cr-contaminated samples in comparison with that in the control samples. The changes in fungal community may be caused by the direct toxic effects on fungi by high concentration of Cr and the significant change in soil properties resulting from Cr contamination. Among all the Cr fractions, organic matter-bound Cr and exchangeable Cr showed significant effects on the fungal community and organic matter also showed a significant effect on soil fungal community.


