J. Cent. South Univ. Technol. (2008) 15: 845-852
DOI: 10.1007/s11771-008-0156-y![]()
Elitism-based immune genetic algorithm and its application to
optimization of complex multi-modal functions
TAN Guan-zheng(谭冠政), ZHOU Dai-ming(周代明), JIANG Bin(江 斌), DIOUBATE Mamady I
(School of Information Science and Engineering, Central South University, Changsha 410083, China)
Abstract:
A novel immune genetic algorithm with the elitist selection and elitist crossover was proposed, which is called the immune genetic algorithm with the elitism (IGAE). In IGAE, the new methods for computing antibody similarity, expected reproduction probability, and clonal selection probability were given. IGAE has three features. The first is that the similarities of two antibodies in structure and quality are all defined in the form of percentage, which helps to describe the similarity of two antibodies more accurately and to reduce the computational burden effectively. The second is that with the elitist selection and elitist crossover strategy IGAE is able to find the globally optimal solution of a given problem. The third is that the formula of expected reproduction probability of antibody can be adjusted through a parameter b, which helps to balance the population diversity and the convergence speed of IGAE so that IGAE can find the globally optimal solution of a given problem more rapidly. Two different complex multi-modal functions were selected to test the validity of IGAE. The experimental results show that IGAE can find the globally maximum/minimum values of the two functions rapidly. The experimental results also confirm that IGAE is of better performance in convergence speed, solution variation behavior, and computational efficiency compared with the canonical genetic algorithm with the elitism and the immune genetic algorithm with the information entropy and elitism.
Key words:
1 Introduction
The biological immune system is a highly parallel and distributed adaptive system, which exists in the body of a vertebrate and is used to recognize and eliminate dangerous foreign antigens to keep the body from damage. In the immune system, foreign bacteria, viruses are regarded as non-self substances and called antigens, and the substances responsible for recognizing and eliminating these non-self substances are called antibodies.
Like the genetic system and brain-nervous system, the immune system is also an information system of human body and has the abilities such as pattern recognition, parallel information processing, memory, associative retrieval, and learning. It is because of these excellent abilities that many experts in the domains of engineering are interested in the immune system. From the requirements of given problems, researchers have developed a number of artificial immune systems (AISs) and artificial immune algorithms (AIAs) through extracting some features from the biological immune system to solve complex engineering problems. So far, three main immunological principles have been adopted in AISs and AIAs, including the immune network theory, the negative selection mechanisms, and the clonal selection principles[1]. Up to now, AISs and AIAs have been applied successfully to many fields of science and engineering, especially in the field of computer science, such as virus elimination and intrusion detection[2], multi-modal function optimization[3], multi-objective optimization[4], e-mail classification[5], data mining[6], machine learning and pattern recognition[7-8], robot control[9-12], medical diagnosis system[13], distributed defense in modern data networks[14], remote-sensing imagery[15].
Genetic algorithms (GAs) are the most widely used evolutionary algorithms, but they are difficult to maintain a good population diversity in an evolutionary process, which often leads GAs to premature convergence. Aiming at this demerit of GAs, CHUN et al[16] proposed an immune genetic algorithm (IGA) by combining the immune network theory and the genetic algorithm, in which the regulative mechanism of antibody diversity was introduced to the genetic algorithm for the first time. Later, FUKUDA et al[3] proposed the information entropy-based IGA by introducing the information entropy to the computation of antibody concentration and similarity. IGAs are able to adjust the selection pressure of GAs effectively and keep a population in good diversity. But, the current IGAs have some demerits, including easily getting trapped in local optima, a slow running speed, and a slow convergence speed.
In this work, a novel immune genetic algorithm was presented by combining the merits of current AIAs and GAs. In this algorithm, the new computational methods for the immunological quantities, including the antibody similarity, expected reproduction probability, and clonal selection probability, were given, and the strategies of elitist selection and elitist crossover were also adopted. This algorithm is called the immune genetic algorithm with the elitism (IGAE). In order to test the validity of IGAE, two different complex multi-modal functions, that is, the needle-in-a-haystack function and the Rastringin’s function, were selected for simulation experiments. In addition, the canonical genetic algorithm with the elitism (CGAE) and the immune genetic algorithm with the information entropy and elitism (IGAIEE) were selected for comparison with IGAE to evaluate if IGAE has better performance.
2 Important definitions of immune genetic algorithms (IGAs)
Similar to GAs, the algorithmic structure of an IGA also includes selection, crossover and mutation operations. However, the population updating strategy of an IGA is different from that of GAs. In an IGA, the selection probability of an individual is related to its fitness and its concentration.
2.1 Important definitions
Assuming that in a population each antibody can be represented by a one-dimensional array with n elements, the definitions of the related immunological quantities are given as follows.
Definition 1 Similarity of antibodies: Assume that v={v1, v2, …, vn} and w={w1, w2, …, wn} are any two antibodies of an antibody population with size m, and their fitness values are denoted by fv and fw, respectively. Assume that e 1 and e 2 are two small positive constants. If
1-ε1≤Ss(v, w)=![]() ≤1+ε1 (1)
≤1+ε1 (1)
1-ε2≤Qs(v, w)=![]() ≤1+ε2 (2)
≤1+ε2 (2)
are satisfied, antibodies v and w are said to be similar to each other, where Ss(v, w) and Qs(v, w) are the indexes reflecting the similarities of antibodies v and w in structure and in quality, respectively; e 1 and e 2 are also called the similarity thresholds. The structure of an antibody means its n elements; the quality of an antibody means its fitness, and the larger the fitness, the better the quality.
Definition 2 Concentration of antibody: In an antibody population with size m, the number of antibodies similar to an antibody k (k=1, 2, …, m), including antibody k itself, is called the concentration of antibody k, which is denoted by ck.
Definition 3 Expected reproduction probability of antibody: In an antibody population with size m, the expected reproduction probability ek of antibody k (k=1, 2, …, m) is defined as
![]() (3)
(3)
where fk and ck are the fitness and concentration of antibody k, respectively; b is a parameter which controls the relative importance of antibody concentration versus antibody fitness in ek. Adjusting b helps to balance the population diversity and the algorithm convergence speed in an evolutionary process.
Definition 4 Selection probability: In an antibody population with size m, the selection probability Psk of antibody k (k=1, 2, …, m) is defined as
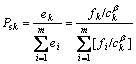 (4)
(4)
Definition 5 Diversity of population: For an antibody population with size m, the diversity index Idiv of the population is defined as
Idiv=![]() (5)
(5)
2.2 Adaptability of new similarity definition
Eqns.(1) and (2) form a new definition method for the similarity of antibodies. The new definition method reflects the two important features of antibody similarity, i.e. the similarity in structure and the similarity in quality. It can be proved that the new definition has the smallest computational complexity compared with the currently used antibody similarity definitions based on the information entropy[3] and the Euclidean distance[17].
However, there is a precondition when using Eqn.(1), that is, the value range of each element of an antibody must be absolutely over a positive or a negative interval, and does not include zero. The precondition of using Eqn.(2) is that the fitness of an antibody is not equal to zero.
In general, for some optimization problems, the precondition of Eqn.(1) is not satisfied. In order to overcome this drawback, for an element whose value range comprises both positive numbers and negative numbers, we can translate its coordinate axis leftward or rightward by a distance to convert its value range into a new one that comprises only positive or negative numbers, and does not include zero.
When translating a coordinate axis, the following suggestions should be considered. Firstly, when using Eqn.(1), in order to reduce the distortion of the ratio vi/wi (i=1, 2, …, n), the translation distance of the coordinate axis of element i should be as small as possible. Secondly, the new value range of each element can be selected whether over a positive or a negative interval, in principle the interval obtained through a smaller-distance translation is better. For example, Fig.1 shows the initial value range and two possible new value ranges of some element xk, where the new one over the negative interval is better. In Fig.1, after the translation, xk is replaced by ![]() Thirdly, because the initial value ranges of all elements may have different widths, the translation distances of their coordinate axes may be different. Fourthly, the value ranges of all elements do not need to be uniformly positive or uniformly negative.
Thirdly, because the initial value ranges of all elements may have different widths, the translation distances of their coordinate axes may be different. Fourthly, the value ranges of all elements do not need to be uniformly positive or uniformly negative.

Fig.1 Translation method of initial value range of element xk
For antibodies v={v1, v2, …, vn} and w={w1, w2, …, wn}, assuming that after the coordinate axes of their elements are translated by a distance l1, l2, …, ln respectively, the new value ranges of their elements are all absolutely over a positive or a negative interval and does not include zero. Set l={l1, l2, …, ln}, where l is called the coordinate translation array.
After translation, antibodies v and w are denoted by ![]() and
and ![]() respectively. The relationships between
respectively. The relationships between ![]() and v,
and v, ![]() and w can be expressed as follows:
and w can be expressed as follows:
![]() (6)
(6)
![]() (7)
(7)
![]() and
and ![]() can be used to replace antibodies v and w in Eqn.(1) for judging their similarity.
can be used to replace antibodies v and w in Eqn.(1) for judging their similarity.
3.1 Elitist selection strategy
A canonical genetic algorithm (CGA) with mutation probability Pm?(0, 1), crossover probability Pc?[0, 1], and the proportional selection never converges to the global optimum regardless of the initialization, crossover operator, and objective function[18]. The reason is that because of the operations of selection, crossover and mutation, the best individual in the population of the current generation is often lost and cannot get into the next generation. In order to prevent the occurrence of this case, de JONG proposed the “elitist selection” or “elitism” strategy[19]. The idea of the strategy is that the best individual found over time is always maintained in the population, either before or after selection. The best individual is also called elitist, which has the largest fitness and the best gene structure.
With the elitist selection strategy, the best individual found over time is no longer lost and destroyed by the selection, crossover and mutation operations in the evolutionary process of the population. This strategy has a great effect on improving the global convergence ability of a CGA. Using the homogeneous finite Markov chain theory, RUDOLPH[18] has proved that the CGA with the elitist selection strategy is able to converge to the global optimum.
3.2 Elitist crossover strategy
The elitist crossover is a new crossover strategy, which means that in the population of each generation, in addition to performing the traditional crossover operations, some individuals are selected to crossover with the elitist using the pre-specified elitist crossover probability to improve the gene structures of the selected individuals[17].
The principle of the strategy can be described as follows: a random number d is generated within [0, 1] for each individual ai(t) in the tth generation, where i=1, 2, …, m, and m is the population size. If d<Pkc, the pre-specified elitist crossover probability, ai(t) is selected to crossover with the elitist individual a*(t) using the specified crossover strategy to generate a pair of offspring a′i(t) and a*′(t). Then, ai(t) in the population is replaced by a′i(t), and a*′(t) is discarded.
The elitist crossover strategy provides more crossover opportunities for the elitist individual than for other individuals, which is advantageous to increasing the excellent schemata of the population. But, the elitist crossover probability Pkc should be small enough, otherwise the diversity of the population will be destroyed because of the excessive inbreeding.
The combinative use of the traditional crossover and elitist crossover is helpful both to maintaining the good diversity of a population and to improving the quality of the population rapidly.
4 IGAE immune genetic algorithm
Based on the above-mentioned computational methods of the immunological quantities, the elitist selection strategy and the elitist crossover strategy, the proposed IGAE algorithm can be given as follows.
Step 1 Randomly generate m antibodies to constitute the initial population of antibodies.
Step 2 Determine the fitness of each antibody and save the elitist antibody with the largest fitness into a specified variable.
Step 3 If the population is the population of the first generation, go to Step 6; otherwise, continue.
Step 4 Compute the fitness fk of each antibody k (k=1, 2, …, m). If there is not an antibody in the population of this generation which has the same fitness as the elitist antibody, add a copy of the elitist antibody saved in the specified variable into the population, and delete the antibody with the smallest fitness from the population; otherwise, continue.
Step 5 If the fitness of the antibody with the largest fitness in the population of this generation is larger than that of the elitist antibody saved in the specified variable, take the antibody as the new elitist antibody and replace the old one saved in the specified variable with the new one; otherwise, continue.
Step 6 Compute the concentration of each antibody by using Eqns.(1) and (2).
Step 7 Compute the expected reproduction probability of each antibody by adopting Eqn.(3); compute the selection probability of each antibody by using Eqn.(4); perform a selection operation on each antibody by using Roulette Wheel Selection method according to its selection probability.
Step 8 Perform crossover operations on the population using the crossover probability Pc?[0, 1].
Step 9 Perform elitist crossover operations on the population using a small crossover probability Pkc.
Step 10 Perform mutation operations on the population using the mutation probability Pm?(0, 1).
Step 11 Examine if the prescribed stopping criterion is satisfied. If it is satisfied, output the optimal solution corresponding to the elitist antibody, and then stop; otherwise, return to Step 4 to continue the iterative repetitions.
In the late stage of evolutionary process, since the quality of each antibody has been improved greatly, in order to keep the population size of each generation unchanged, in Step 4 antibody with higher concentration can be deleted from the population to maintain the population in a good diversity.
5 Experiments and analysis5.1 Testing functions
The discontinuous needle-in-a-haystack function and the Rastringin’s function were selected as the testing functions to evaluate the performance of the proposed IGAE algorithm, and IGAE was used to search for the globally maximum or minimum value of the two functions.
The form of the needle-in-a-haystack function is
 (8)
(8)
0≤x<5.12, -5.12≤y≤5.12
where a=3.0, b=0.05. This is a trickish function for GAs and GAs are easily allured into its local optimal solutions. When x=0 and y=0, the function gets its globally maximum value of f = 3 690. Let 0-=lim x, the point (x=0-, y=0) is the trickish attractor of the function, at which the value of the function approaches to 3 600. In this work, this function is simply denoted by F1.
The form of the Rastringin’s function with N independent variables is
![]()
-5.12≤xi≤5.12, i=1, 2, …, N (9)
where x ={x1, x2, …, xN }. At point x={x1, x2, …, xN}={0, 0, …, 0}, the function gets its globally minimum value, i.e. zero. This is a complex multi-modal function and has an extremely large search space and numbers of local minimum values. It is very difficult to search for its globally minimum value. In this work, A and N are set to 10, respectively, and the function is simply denoted by F2.
Figs.2 and 3 show the three-dimensional figures of F1 and F2 (here F2 has only two independent variables), respectively. From the figures, it can be seen that F1 has several discontinuous and isolated peaks, whereas F2 has large numbers of peaks.
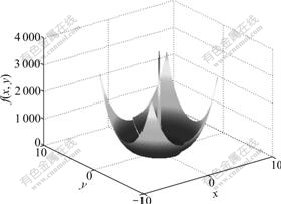
Fig.2 Three-dimensional figure of F1
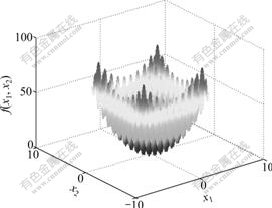
Fig.3 Three-dimensional figure of F2 with only two independent variables
5.2 Effects of parameters e1, e2 and b on performance of IGAE
When using IGAE to search for the globally maximum value of F1, the parameters are specified as: the population size m=180; the maximum number of generations NC=500; the length of a chromosome Lch=60 bit, where variables x and y occupy 30 bit respectively; the crossover probability Pc=0.6, using the two-point crossover strategy; the elitist crossover probability Pkc=0.2; for the mutation probability Pm, if the fitness of an individual is smaller than 95% of that of the elitist individual, then Pm=0.05, otherwise Pm=0.005. If the maximum function value produced by IGAE is smaller than 3 689.999 9, then IGAE fails to converge to the globally maximum value 3 690 of F1 within the number of generations NC.
When using IGAE to search for the globally minimum value of F2, the parameters are specified as: the population size m=180; the maximum number of generations NC=10 000; the length of a chromosome Lch= 240 bit, where each variable xi occupies 24 bit, i=1, 2, …, N and N=10; the crossover probability Pc=0.6, using the ten-point crossover strategy; the elitist crossover probability Pkc=0.2; for the mutation probability Pm, if the fitness of an individual is smaller than 95% of that of the elitist individual, then Pm=0.01, otherwise Pm=0.001. If the minimum function value produced by IGAE is larger than 0.000 1, then IGAE fails to converge to the globally minimum value of F2, i.e. zero, within the number of generations NC.
5.2.1 Effects of e 1 and e 2 on performance of IGAE
For F1, the coordinate translation array was selected as l={l1, l2}={10.24, 10.24}; for F2, the coordinate translation array was selected as l={l1, l2, …, l10}= {10.24, 10.24, …, 10.24}. In order to investigate the effects of e1 and e2 on the performance of IGAE, for F1 and F2 30 random experiments were performed respectively to search for their globally maximum or minimum values using a fixed b (b =1) and different combinations of e1 and e2. The experiment results are shown in Tables 1 and 2.
Table 1 Effects of e1 and e2 on performance of IGAE (Function F1)

Table 2 Effects of e1 and e2 on performance of IGAE (Function F 2)

From Tables 1 and 2, it can be seen that e1 and e2 have a great influence on the search results and convergence speed of IGAE. When e1=0.10, e2=0.20, IGAE can find the globally maximum value (3 690 ) of F1 in the shortest time. When e1=0.30, e2=0.30, the solution produced by IGAE for F2 is the best.
5.2.2 Effect of b on performance of IGAE
For F1, let e1=0.10, e2=0.20, and l={l1, l2}={10.24, 10.24}; for F2, let e1=0.30, e2=0.30, and l={l1, l2, …, l10}={10.24, 10.24, …, 10.24}. In order to investigate the effect of b on the performance of IGAE, for F1 and F2 30 random experiments were also performed respectively to search for their globally maximum or minimum values using different b values. The results are shown in Tables 3 and 4.
Table 3 Effect of b on performance of IGAE (Function F 1)

Table 4 Effect of b on performance of IGAE (Function F 2)

Tables 3 and 4 show that b has a great influence on the search results and convergence speed of IGAE. It can be seen from Tables 3 and 4 that when b =1, IGAE can find the globally maximum value of F1 and the globally minimum value of F2 in the shortest time.
5.3 Comparison of IGAE with other evolutionary algorithms
In order to evaluate the performance of IGAE, as the comparison objects, the canonical genetic algorithm with the elitism (CGAE) and the immune genetic algorithm with the information entropy and elitism (IGAIEE) were used to search for the globally maximum or minimum values of function F1 and F2. The CGAE was from the method in Ref.[20] and the IGAIEE was from the methods in Refs.[3, 19].
When optimizing F1, the parameters of the three algorithms are specified as: the population size m=180; the maximum number of generations NC=2 000; the length of a chromosome Lch=60 bit, where the variables x and y occupy 30 bit respectively; the crossover probability Pc=0.6, using the two-point crossover strategy; the elitist crossover probability Pkc=0.2; for the mutation probability Pm, if the fitness of an individual is smaller than 0.95% that of the elitist individual, then Pm= 0.05, otherwise Pm=0.005. Other parameters are specified as: in IGAIEE, the threshold of antibody similarity Tac=0.7; in IGAE, e1=0.10, e 2=0.20, b=1, and l={10.24, 10.24}.
When optimizing F2, the parameters of the three algorithms are specified as: the population size m=180; the maximum number of generations NC=10 000; the length of a chromosome Lch=240 bit, where each variable xi occupies 24 bit, i=1, 2, …, N and N=10; the crossover probability Pc=0.6 using the ten-point crossover strategy; the elitist crossover probability Pkc=0.2; for the mutation probability Pm, if the fitness of an individual is smaller than 95% of that of the elitist individual, then Pm=0.01, otherwise Pm=0.001. Other parameters are specified as: in IGAIEE, the threshold of antibody similarity Tac=0.7; in IGAE, e1=0.30, e2=0.30, b=1, and l={l1, l2, …, l10}={10.24, 10.24, …, 10.24}.
5.3.1 Comparison of convergence speed
Using the same initial population generated randomly, the experiments were performed using each of the three algorithms IGAE, CGAE, and IGAIEE to search for the globally maximum value of F1 (F1, max) and the globally minimum value of F2 (F2, min). The results are shown in Figs.4 and 5. It can be seen that the convergence speed of IGAE is faster than those of CGAE and IGAIEE.
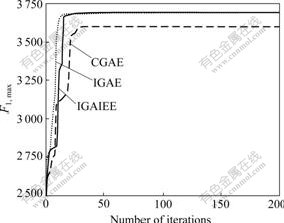
Fig.4 Convergence tendency of the maximum function value of F1 in one experiment
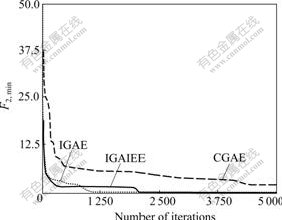
Fig.5 Convergence tendency of the minimum function value of F2 in one experiment
5.3.2 Comparison of solution variation behavior
In order to compare the solution variation behavior of the three algorithms in many experiments, 30 random experiments were performed using each of the three algorithms to search for the globally maximum/minimum values of functions F1 and F2. In each experiment, the initial population was generated randomly. The following sample standard deviation d was used as the evaluation index of solution variation behavior:
![]() (10)
(10)
where Ji is the optimal function value obtained in the ith experiment and ![]() is the average value of the optimal function values obtained in N experiments. Table 5 shows the d value of each algorithm.
is the average value of the optimal function values obtained in N experiments. Table 5 shows the d value of each algorithm.
Table 5 Comparison of d values of three algorithms in 30 random experiments
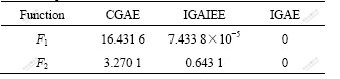
For both F1 and F2, the d value of the IGAE is the smallest among the three algorithms, which means that when repeatedly performing many experiments the optimal values generated by IGAE have the smallest variation and the best stability.
5.3.3 Comparison of computational efficiency
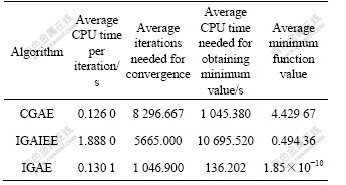
30 random experiments were performed using each of the three algorithms to search for the maximum value of F1 and the minimum value of F2. In each experiment the initial population was generated randomly. Tables 6 and 7 show the computational efficiency data of the three algorithms. In these experiments, a computer with 1.85 GHz CPU (AMD2500+) and 256 MB RAM was used, and the operating system was the Windows XP. It can be seen from these statistical data that although the average CPU time of IGAE needed for completing an iteration is not the smallest, because its average number of iterations needed for convergence is the smallest, its average CPU time needed for obtaining the globally maximum/ minimum value is also the smallest among the three algorithms.
Table 6 Comparison of computational efficiency of three algorithms (Function F1)
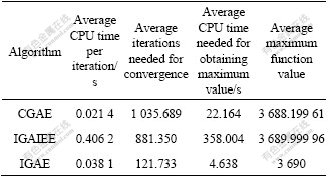
Table 7 Comparison of computational efficiency of three algorithms (Function F2)
1) A novel immune genetic algorithm called IGAE is proposed based on the elitist selection strategy, elitist crossover strategy, and new methods for computing the antibody similarity, expected reproduction probability, and clonal selection probability. The simulation experiments demonstrate that IGAE has global convergence ability.
2) Compared with the canonical genetic algorithm with the elitism and the immune genetic algorithm with the information entropy and elitism, the simulation experiments demonstrate that IGAE has better performance in convergence speed, solution variation behavior, and computational efficiency.
3) Adjusting the immunological quantity control parameters e1, e2 and b is helpful to balancing the population diversity and the algorithmic convergence speed in an evolutionary process, which enables IGAE to converge to the globally optimal solution of a given problem rapidly.
References[1] TIMMIS J, HONE A, STIBOR T, CLARK E. Theoretical advances in artificial immune systems [J]. Theoretical Computer Science, 2008, 403(1): 11-32.
[2] DASGUPTA D, GONZALEZ F. Enhancing computer security with smart technology [M]. New York: Auerbach Publications, 2005: 165-208.
[3] FUKUDA T, MORI K, TSUKIYAMA M. Artificial immune systems and their applications [M]. Heidelberg: Springer-Verlag Press, 1999: 210-220.
[4] TAN K C, GOH C K, MAMUN A A, EI E Z. An evolutionary artificial immune system for multi-objective optimization [J]. European Journal of Operational Research,2008, 187(2): 371-392.
[5] SECKER A, FREITAS A, TIMMIS J. AISEC: An artificial immune system for E-mail classification [C]// Proceedings of the 2003 IEEE Congress on Evolutionary Computation. Piscataway, New Jersey: IEEE Press, 2003: 131-139.
[6] FREITAS A A, TIMMIS J. Revisiting the foundations of artificial immune systems for data mining [J]. IEEE Transactions on Evolutionary Computation, 2007, 11(4): 521-540.
[7] GLICKMAN M, BALTHROP J, FORREST S. A machine learning evaluation of an artificial immune system [J]. Journal of Evolutionary Computation, 2005, 13(2): 179-212.
[8] de CASTRO L N, von ZUBEN F J. Learning and optimization using the clonal selection principle [J]. IEEE Transactions on Evolutionary Computation, 2002, 6(3): 239-251.
[9] DIOUBATE M I, TAN G Z, TOUR M L. An artificial immune system-based multi-agent model and its application to robot cooperation problem [C]// Proceedings of the 7th World Congress on Intelligent Control and Automation. Piscataway, New Jersey: IEEE Press, 2008: 3033-3039.
[10] DIOUBATE M I, TAN G Z. Immune evolutionary programming- based locomotion control of autonomous mobile robot [C]// Proceedings of the 7th World Congress on Intelligent Control and Automation. Piscataway, New Jersey: IEEE Press, 2008: 8990-8996.
[11] DIOUBATE M I, TAN G Z. Robot goal-discovering problems-based adapted idiotypic immune network [C]// Proceedings of The 2008 International Conference on Genetic and Evolutionary Methods (GEM’08). New York: CSREA Press, 2008: 203-211.
[12] NASSER H A, SELIM G, BADE A. A fuzzy-immune algorithm for autonomous robot navigation [J]. Computing and Information Systems, 2007, 11(3): 12-22.
[13] GUNES S, POLAT K. Computer aided medical diagnosis system based on principal component analysis and artificial immune recognition system classifier algorithm [J]. Expert Systems with Applications, 2008, 34(1): 773-779.
[14] SWIMMER M. Using the danger model of immune systems for distributed defense in modern data networks [J]. Computer Networks, 2007, 51(5): 1315-1333.
[15] ZHONG Yan-fei, ZHANG Liang-pei, GONG Jian-yai, LI Ping-xiang. A supervised artificial immune classifier for remote-sensing imagery [J]. IEEE Transactions on Geoscience and Remote Sensing, 2007, 45(12): 3957-3966.
[16] CHUN J S, JUNG H K, HAHN S Y. A study on comparison of optimization performance between immune algorithm and other heuristic algorithms [J]. IEEE Transactions on Magnetics, 1998, 34(5): 2972-2975.
[17] ZHENG Ri-rong, MAO Zong-yuan, LUO Xin-xian. Study of immune algorithm based on Euclidean distance and king-crossover [J]. Control and Decision, 2005, 20(2): 161-164, 169. (in Chinese)
[18] RUDOLPH G. Convergence analysis of canonical genetic algorithms [J]. IEEE Transactions on Neural Networks, 1994, 5(1): 96-101.
[19] de JONG K A. An analysis of the behavior of a class of genetic adaptive systems [D]. Ann Arbor: University of Michigan, 1975: 25-97.
[20] BHANDARI D, MURTHY C A, PAL S K. Genetic algorithm with elitist model and its convergence [J]. International Journal of Pattern Recognition and Artificial Intelligence, 1996, 10(6): 731-747.
Foundation item: Project(50275150) supported by the National Natural Science Foundation of China; Projects(20040533035, 20070533131) supported by the National Research Foundation for the Doctoral Program of Higher Education of China
Received date: 2008-03-05; Accepted date: 2008-04-12
Corresponding author: TAN Guan-zheng, Professor; Tel: +86-731-8716259; E-mail: tgz@mail.csu.edu.cn
(Edited by CHEN Wei-ping)
Abstract: A novel immune genetic algorithm with the elitist selection and elitist crossover was proposed, which is called the immune genetic algorithm with the elitism (IGAE). In IGAE, the new methods for computing antibody similarity, expected reproduction probability, and clonal selection probability were given. IGAE has three features. The first is that the similarities of two antibodies in structure and quality are all defined in the form of percentage, which helps to describe the similarity of two antibodies more accurately and to reduce the computational burden effectively. The second is that with the elitist selection and elitist crossover strategy IGAE is able to find the globally optimal solution of a given problem. The third is that the formula of expected reproduction probability of antibody can be adjusted through a parameter b, which helps to balance the population diversity and the convergence speed of IGAE so that IGAE can find the globally optimal solution of a given problem more rapidly. Two different complex multi-modal functions were selected to test the validity of IGAE. The experimental results show that IGAE can find the globally maximum/minimum values of the two functions rapidly. The experimental results also confirm that IGAE is of better performance in convergence speed, solution variation behavior, and computational efficiency compared with the canonical genetic algorithm with the elitism and the immune genetic algorithm with the information entropy and elitism.


