- Abstract:
- 1 Introduction▲
- 2 Materials and methods▲
- 2.1 Samples and culture media
- 2.2 Screening and identification of chitinase- producing strain
- 2.3 Inducibility
- 2.4 Production and purification of chitinase secreted by strain CS-01
- 2.5 Electrophoresis
- 2.6 Protein determination
- 2.7 Determination of chitinase activity
- 2.8 Determination of chitinase stability
- 2.9 Nucleotide sequence accession number
- 3 Results and discussion▲
- 4 Conclusions▲
- References
- Figure
- Fig.1 Morphological features of strain CS-01: (a) Colony formed on solidified Czapek-Dox medium for 4 d; (b) Colony formed on solidified selected medium for 4 d; (c), (d) Microscopic morphologies
- Fig.2 Phylogenetic tree from ITS domain of 28S rDNA gene sequence of CS-01
- Fig.3 Time courses of chitinase activity of A. fumigatus CS-01 cultivated by different carbon sources
- Fig.4 Time courses of reducing sugar and chitinase activity present in culture supernatant of A. fumigatus CS-01 cultured by mixed carbon source containing 0.5% glucose and 0.5% colloidal chitin
- Fig.5 Native-PAGE analysis results of purified product (Lane 1, relative molecular mass maker; Lane 2, sample) (a), and relative molecular mass determination of chitinase by SDS-PAGE (Lane 1, maker proteins; Lane 2, purified chitinase) (b)
- Fig.6 Effect of pH on activity and stability of chitinase
- Fig.7 Effect of temperature on activity and stability of chitinase
J. Cent. South Univ. Technol. (2009) 16: 0552-0557
DOI: 10.1007/s11771-009-0092-5
![]()
Purification and characterization of extracellular chitinase from a novel strain Aspergillus fumigatus CS-01
XIA Jin-lan(夏金兰)1, 2, XIONG Jing(熊 晶)1, 2, XU Tao(徐 涛)1, 2, ZHANG Cheng-gui(张成桂)1, 2,
ZHANG Rui-yong(张瑞永)1, 2, ZHANG Qian(张 倩)1, 2, WU Shun(武 顺)1, 2, QIU Guan-zhou(邱冠周)1, 2
(1. Key Laboratory of Biometallurgy of Ministry of Education, Central South University, Changsha 410083, China;
2. School of Resources Processing and Bioengineering, Central South University, Changsha 410083, China)
Abstract:
Twelve samples derived from different locations in south central area of China are treated by enrichment and spread-plate technique for initial screening. Seven chitinase-producing strains are isolated. The chitinase present in the culture supernatant of strain CS-01 possesses the maximum activity of 0.118 U/mL. Analysis of the morphological feature and the ITS rDNA sequence reveals that strain CS-01 belongs to Aspergillus fumigatus. Production of the chitinase is regulated by a inducible way and the maximum activity appears at 36 h in colloidal chitin culture. Purification of the chitinase is carried out by salting out, gel filtrate chromatography and anion exchange chromatography sequentially. Native-PAGE and SDS-PAGE indicate that the chitinase from A. fumigatus CS-01 is a monomer with the relative molecular mass estimated to be 4.50×104. Its maximum activity appears at pH 5 and 55 ℃. The chitinase is stable at pH 4.0-7.5 and below 45 ℃.
Key words:
Aspergillus fumigatus; chitinase; purification;
1 Introduction
Chitin, a linear polymer of β-1, 4-N-acetylglucose- amine units with abundance next to cellulose, is the most abundant natural amino polysaccharide [1]. It is widely distributed as a structural component of crustaceans, insects, as well as a component of the cell walls of most fungi and some algae [2]. Previous reports indicated that these natural polymers and their degraded products have excellent properties such as biocompatibility, biodegradability, non-toxicity, and adsorption properties [3] so as to cause strong interest from worldwide researchers in its application potentials [4].
Although chemical methods and physical methods have been industrialized, they have several disadvantages of high cost, low yield and no specificity, even environmental pollution [5-7]. More concern is focused on enzymatic methods due to their regioselective depolymerization under mild conditions [3]. Enzymatic degradation can be performed by chitinase, chitosanase, glucanase, etc [8]. Previous studies revealed that chitinase has a variety of application potentials (e.g. preparation of fungi protoplast, cytochemistry location, antifungal agent and transgenetic plant) [9-11]. In order to realize bioconversion of chitinous material and find out excellent chitinase with versatile abilities of heat
tolerance, high specificity and stability, thorough studies on widespread chitinase are indispensable.
In this work, we report the screening and identification of chitinolytic strains from soil and water samples, the investigation of the inducible secretion of chitinase from one of the strains, Aspergillus fumigatus CS-01, as well as the purification and characterization of that secreted chitinase.
2 Materials and methods
2.1 Samples and culture media
For the enrichment of chitinolytic microorganisms, soil and water from streams and shrimp farms are collected at 12 different locations in south central area of China. Colloidal chitin is prepared from powdered chitin (Sanland) by the method of XIAO et al [12]. Enrichment medium (pH 6.0) contains 1% colloidal chitin, 0.5% peptone, 0.05% MgSO4?7H2O, 0.24% KH2PO4 and 0.06% K2HPO4. Selective medium (pH 6.0) consists of 1% colloidal chitin, 0.5% (NH4)2SO4, 0.24% KH2PO4, 0.06% K2HPO4?3H2O and 0.05% MgSO4·7H2O. Solidified selective medium is obtained by adding 1.5% agar into selective medium. Czapek-Dox medium contains 0.2% NaNO3, 0.1% K2HPO4?3H2O, 0.05% KCl, 0.05% MgSO4?7H2O, 0.001% FeSO4?7H2O and 2%
sucrose. Solidified Czapek-Dox medium is obtained by adding 1.5% of agar into Czapek-Dox medium. Colloidal chitin agar slant (pH 6.0) consists of 1% colloidal chitin, 0.05% MgSO4?7H2O, 0.5% (NH4)2SO4 and 1.5% agar. General culture medium (pH 6.0) contains 1% colloidal chitin, 0.05% MgSO4?7H2O and 0.5% (NH4)2SO4.
2.2 Screening and identification of chitinase- producing strain
The 12 samples are respectively added into the sterile enrichment medium, followed by incubation under aerobic condition at 35 ℃ for 24 h. The enrichment culture is gradually diluted and then spread on solidified selective medium, and incubated at 35 ℃ for 5 d. Colonies with clear zone against the creamy background are regarded as chitinase-producing and respread until pure cultures are obtained. Equal inoculum density of the isolated strains is subcultured at 35 ℃ and 180 r/min for 36 h, and then collected for measurement of chitinase activity. The strain with highest chitinase activity present in supernatant is maintained on colloidal chitin agar slant, and used for further study.
Identification of strain CS-01 is performed by morphological feature and ITS rDNA sequence analysis [13-15]. Colonial feature and hypha modality are observed [13-14]. The ITS rDNA sequence of the strain CS-01 is amplified by PCR using two oligonucleotide primers: 5′-GGTCCGTGTTTCAAGAGC-3′ and 5′-GCA TATCAATAAGCGGAGGAAAAG-3′. The PCR reactions are initiated at 95 ℃ (3 min), followed by 30 cycles at 94 ℃ (30 s), annealing at 55 ℃ (30 s), 72 ℃ (1 min), and ended with incubation at 72 ℃ for at least 5 min. The interesting fragment from PCR product separated by 0.6% agarose gel is excised from the gel and purified with Rapid Recovery Kit (Gel), followed by DNA sequencing (Shanghai Sangon). Homological sequence alignment of the ITS rDNA sequence with other origin from Genebank is performed by using ClustalX 1.8 software, and then phylogenetic tree is constructed with MEGA3.1 software.
2.3 Inducibility
To investigate the required condition for chitinase production from strain CS-01, the carbon source respectively served by 1% glucose, 1% colloidal chitin, and mixed carbon source containing 0.5% glucose and 0.5% colloidal chitin, is supplemented with the medium (pH 6.0) containing 0.05% MgSO4·7H2O and 0.5% (NH4)2SO4. Equal inoculum density is inoculated and cultivated at 35 ℃ on a rotary shaker (180 r/min). Chitinase activity is measured at 12 h intervals for up to 60 h.
2.4 Production and purification of chitinase secreted by strain CS-01
Strain CS-01, identified as Aspergillus fumigatus, is utilized to produce chitinase. According to 10% inoculum density, its diluted spores (106 mL-1) are inoculated into the general culture medium, and cultivated at 35 ℃ for 36 h on a rotary shaker (180 r/min). The culture filtrate for purification is obtained by suction method. Unless otherwise specified, all purification steps are carried out at 4 ℃.
(1) Ammonium sulfate precipitation
The culture filtrate is fractionated with ammonium sulfate of 80% saturation. After stay overnight, the protein precipitate is obtained by centrifugation at 12 000 r/min for 15 min. The precipitate is dissolved in small volumes of 0.05 mol/L Tris buffer (pH 7.5).
(2) Gel filtration chromatography
Sephadex G-75 (Pharmacia) is prepared by the method of the Pharmacia Company. Sephadex G-75 exposed in 0.05 mol/L Tris buffer (pH 7.5) is incubated in the boiling water for 30 min at 100 ℃ for the column (1.5 cm×60 cm). The column is equilibrated with 0.05 mol/L Tris buffer (pH 7.5) at a constant flow rate of 0.25 mL/min. The concentrated enzyme derived from salting out is applied to the column and eluted with the same buffer. Fractions of 1.5 mL are collected at a flow rate of 0.25 mL/min. Protein profile is monitored by measuring the absorbance at 280 nm. The fractions that possess chitinase activity are pooled, dialyzed overnight against 0.02 mol/L Tris buffer (pH 8.5), and then concentrated by solid polyethylene glycol for next step.
(3) Anion exchange chromatography
DEAE-Sephadex A-25 (Pharmacia) is prepared by the method of the Pharmacia Company. The sample is applied to a DEAE-Sephadex column (1.5 cm×60 cm) equilibrated with 0.02 mol/L Tris buffer (pH 8.5). The enzyme is eluted with a linear gradient of NaCl from 0 to 1 mol/L in 0.02 mol/L Tris buffer (pH 8.5). Fractions of 1.5 mL are collected at a constant flow rate of 0.5 mL/min. The fractions that possess chitinase activity are pooled, dialyzed overnight against sterile distilled water, and then concentrated by freeze-drying. The concentrated enzyme is kept at -20 ℃ until use.
2.5 Electrophoresis
Native-PAGE and SDS-PAGE of the purified sample are respectively performed using a Mini Protean II vertical tank apparatus (JUNYI Co. Ltd, China) [16-18]. Protein bands are stained with Coomassie Brilliant Blue R-250 staining solution, followed by decoloration. Broad-range protein markers (Ferentus) are used for the estimation of protein size.
2.6 Protein determination
Protein concentration is determined by using the Bradford method with bovine serum albumin (Ferentus) as a reference protein [19].
2.7 Determination of chitinase activity
Chitinase activity is determined by measuring the amount of the reducing end group degraded from colloidal chitin [20]. One unit of activity is defined as the amount of enzyme that liberates 1 μmol of reducing sugar per minute.
2.8 Determination of chitinase stability
Chitinase stability is determined by the same way as the chitinase activity except that before determination, the chitinase is incubated at the detected pH or temperature for 30 min.
2.9 Nucleotide sequence accession number
The nucleotide sequence of the ITS rDNA of strain CS-01 is deposited with the GenBank database under accession number EU786151.
3 Results and discussion
3.1 Isolation and screening of chitinase-producing strains
Seven strains form colonies of 0.8-1.2 cm in diameter on the plates, surrounded by clear zones indicating chitinase activity, named CS-01-CS-07. Chitinase activities present in the ferment supernatants of strains CS-01-CS-07 determined by colorimetric method are 0.118, 0.091, 0.076, 0.035, 0.047, 0.051 and 0.085 U/mL, respectively. Strain CS-01 with the highest chitinase activity is thus maintained on colloidal chitin agar slant culture medium and used for further study.
3.2 Identification of strain CS-01
Phenotype identification of strain CS-01 is performed by observation of colonial and hyphae features (Fig.1). Colonial morphology of strain CS-01 on solidified Czapek-Dox medium is observed at different growth stages. Initially, the colony is white and villiform with radial circumference. After cultivated for 2-3 d, the central region of the colony gradually changes from white to light green, then to sap green. After 7 d, several colonies are overlapped and become dark green lawn. Clear zones appear on solidified selective medium after cultivated for 3-4 d. Mycelia morphology of the strain cultured on solidified Czapek-Dox medium for 7 d indicates that mycelia possesses smooth conidiophore and vase-like vesicle; conidiophores are globular and green; and hypha fragments are observed at branch site. All of these are mostly similar to the features of A. fumigatus.
Molecular identification of strain CS-01 is carried out by PCR amplification and sequencing of ITS rDNA region. The amplified product is subject to agar electrophoresis (data not shown). The sequencing result indicates that the amplified ITS rDNA region is 491 bp. In terms of homological sequence alignment and phylogenetic tree construction, strain CS-01 shared the most homology with A. fumigatus NRRL 6113 (Fig.2). So, strain CS-01 could be classified as A. fumigatus.
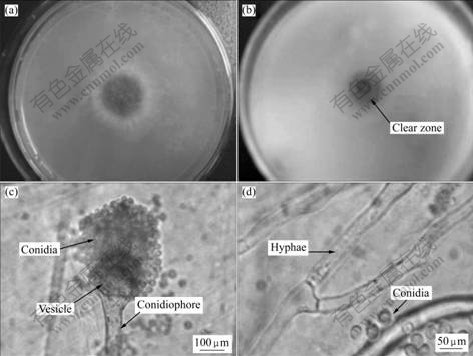
Fig.1 Morphological features of strain CS-01: (a) Colony formed on solidified Czapek-Dox medium for 4 d; (b) Colony formed on solidified selected medium for 4 d; (c), (d) Microscopic morphologies
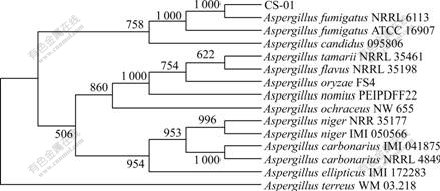
Fig.2 Phylogenetic tree from ITS domain of 28S rDNA gene sequence of CS-01
3.3 Inducibility
A. fumigatus CS-01 is cultivated by different carbon sources. Chitinase activity present in the supernatant of 1% glucose cultivation cannot be detected all the time; while the culture supernatant from 1% colloidal chitin cultivation possesses enzymatic activity after incubated for 12 h and chitinase activity reaches its maximum (0.128 U/mL) at 36 h (Fig.3), which is much more and earlier than that of A. fumigatus TKU003 [17]. A. fumigatus CS-01 cultivated by mixed carbon source shows chitinase activity at late stage of growth (Fig.3). Further investigation is carried out by determining reducing sugar and chitinase activity in the mixed carbon source culture (Fig.4). The above results indicate that the chitinase expression is chitin-inducible.
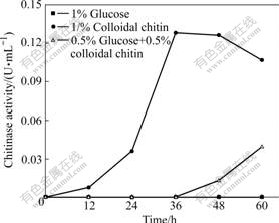
Fig.3 Time courses of chitinase activity of A. fumigatus CS-01 cultivated by different carbon sources
However, there is no consensus about the regulation mechanism of chitinase biosynthesis. As reported, Aspergillus fumigatus NCPF 2140 is regulated by a negative feedback mechanism [18], opposite to the csn gene of Bacillus subtilis 168 [16]. And the chitinase operon of Clostridium paraputrificum can express chitinase under the condition of high concentration of glucose [19]. Thereby, inducibility of chitinase seems to
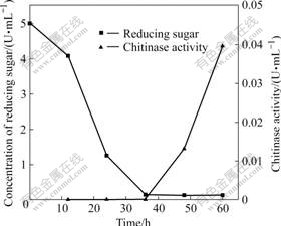
Fig.4 Time courses of reducing sugar and chitinase activity present in culture supernatant of A. fumigatus CS-01 cultured by mixed carbon source containing 0.5% glucose and 0.5% colloidal chitin
conflict with the indispensable role of chitinase in morphogenesis demanded during the fungal life cycle. Whether the chitinases from different strains are encoded by different genes or regulated by different mechanisms is not clear, they need further investigation.
The final specific activity of the purified chitinase is 7.58 U/mg with a purification factor of 16.13. The final amount of the chitinase obtained is 0.31 mg with an overall yield of 1.83%. Quantitative results obtained from the purification steps are shown in Table 1. The purity of the chitinase after purification is validated by Native-PAGE: a single band of chitinase is observed, which indicates that chitinase is highly purified (Fig.5(a)). The purified enzyme is stored at -20 ℃ for further study.
3.5 Characterization of chitinase
Native-PAGE validates the purity of the chitinase and estimates its native relative molecular mass approximately 3.80×104 (Fig.5(a)). SDS-PAGE shows
Table 1 Purification results of chitinase from A. fumigatus CS-01
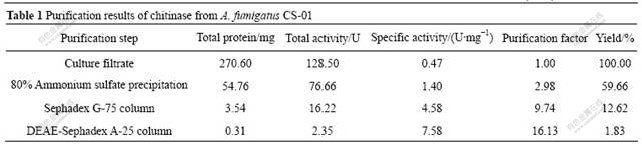
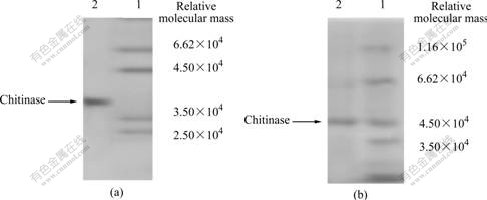
Fig.5 Native-PAGE analysis results of purified product (Lane 1, relative molecular mass maker; Lane 2, sample) (a), and relative molecular mass determination of chitinase by SDS-PAGE (Lane 1, maker proteins; Lane 2, purified chitinase) (b)
that the chitinase from A. fumigatus CS-01 is composed of the same subunit with relative molecular mass of 4.50×104 (Fig.5(b)). Conclusion can be drawn that the chitinase from A. fumigatus CS-01 is monomer with relative molecular mass of 4.50×104.
Effects of pH and temperature on the chitinase activity are investigated by using colloidal chitin as a substrate. Chitinase activity is measured at various pH values (Na2HPO4-citrate buffer, pH 2-6 Na2HPO4- KH2PO4 buffer, pH 6-9) and 35 ℃ for 30 min. The chitinase is stable at pH 4.0-7.5 and its optimum pH is 5.0 (Fig.6). The optimum temperature for the chitinase tested is 55 ℃. The chitinase can maintain over 80% of its initial activity from 30 to 55 ℃ and have 50% of its activity at 65 ℃ (Fig.7).
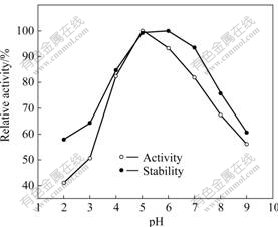
Fig.6 Effect of pH on activity and stability of chitinase
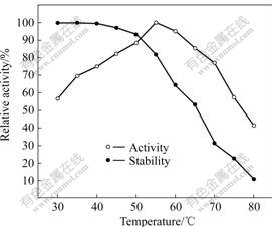
Fig.7 Effect of temperature on activity and stability of chitinase
Compared with other clearly characterized chitinase, CS-01 chitinase possesses more excellently thermo tolerance and less sensitivity to temperature than A. fumigatus YJ-407 origin [20]. The remainder of enzymatic activity after being incubated at 80 ℃ for 30 min is over 10% of the maximum. Moreover, gradually increasing temperature just accompanies a benign decreasing trend of chitinase activity.
4 Conclusions
(1) A new chitin-degrading strain is isolated from south central area of China. According to the physiological features, biochemical properties and gene sequence analysis, the new isolate falls into the species A. fumigatus. We nominate it A. fumigatus CS-01.
(2) Strain CS-01, cultivated by colloidal chitin as sole carbon source, possesses the maximum chitinase activity (0.118 U/mL) present in the culture supernatant, and its expression belongs to inducible system.
(3) The extracellular chitinase from strain CS-01 is isolated, purified and characterized, and it is a monomer with relative molecular mass of 4.50×104, its maximum activity appears at pH 5 and 55 ℃, and it is stable at pH 4.0-7.5 and below 45 ℃.
References
[1] RINAUDO M. Chitin and chitosan: Properties and applications [J]. Progress in Polymer Science, 2006, 31(7): 603-632.
[2] MAJETI N V, RAVI K. A review of chitin and chitosan applications [J]. Reactive and Functional Polymers, 2000, 46(1): 1-27.
[3] PRASHANTH K, THARANATHAN R. Chitin/chitosan: Modifications and their unlimited application potential—An overview [J]. Trends in Food Science & Technology, 2007, 18(3): 117-131.
[4] HIRANO S. Chitin and chitosan as novel biotechnological materials [J]. Polymer International, 1999, 48(8): 732-734.
[5] XING Rong-e, LIU Song, YU Hua-hua, GUO Zhan-yong, WANG Pi-bo, LI Cui-ping, LI Zhi-en, LI Peng-cheng. Salt assisted acid hydrolysis of chitosan to oligomers under microwave irradiation [J]. Carbohydrate Research, 2005, 340(13): 2150-2153.
[6] HIEN N, NAGASAWA N, THAM L, YOSHII F, DANG V H, MITOMO H, MAKUUCHI K, KUME T. Growth-promotion of plants with depolymerized alginates by irradiation [J]. Radiation Physics and Chemistry, 2000, 59(1): 97-101.
[7] ALLAN G, PEYRON M. Molecular weight manipulation of chitosan I: Kinetics of depolymerization by nitrous acid [J]. Carbohydrate Research, 1995, 277(2): 257-272.
[8] PANTALEONE D, YALPANI M, SCOLLAR M. Unusual susceptibility of chitosan to enzymatic hydrolysis [J]. Carbohydrate Research, 1992, 237(1): 325-332.
[9] BOLAR J P, NORELLI J L, WONG K W, HAYES C K, HARMAN G E, ALDWINCKLE H S. Expression of endochitinase from Trichoderma harzianum in transgenic apple increases resistance to apple scab and reduces vigor [J]. Phytopatholigy, 2000, 90(1): 72-77.
[10] ZIMAND G, ELAD Y. Efect of Trichoderma harizianum on Bollrylis cinerea pathogenicity [J]. Phytopatholigy, 1996, 86(5): 945-956.
[11] TRACHUK L, REVINA L, SHEMYAKINA T.Chitinases of bacillus lichenfoumis B-6839: Isolation and properties [J]. Canadian Journal of Micrrobiology, 1996, 42(4): 307-315.
[12] XIAO Xiang, ZHOU Ying, WANG Feng-ping. Isolation and identification of a high-efficient chitin-degrading marine bacterium CB101 and studies on its chitinase system[J]. Acta Oceanologica Sinica, 2003, 25(1): 138-142. (in Chinese)
[13] SHUTTON D, FOTHERQILL A, RINALDI M. Guide to clinically significant fungi [M]. Baltimore: Williams & Wilkins, 1998.
[14] XIA Jin-lan, PENG An-an, HE Huan, YANG Yu, LIU Xue-duan, QIU Guan-zhou. A new strain acidithiobacillus albertensis BY-05 for bioleaching of metal sulfides ores [J]. Transactions of Nonferrous Metals Society of China, 2007, 17(1): 168-175.
[15] INNIS M A, GELFAND D H, SNINSKY J J, WHITE T J. PCR Protocols: A guide to methods and application [M]. San Diego, CA: Academic Press, 1990: 315-322.
[16] RIVAS L A, PARRO V, MORENO-PAZ M. The Bacillus subtilis 168 csn gene encodes a chitosanase with similar properties to a streptomyces enzyme [J]. Microbiology, 2000, 146(1): 2929-2936.
[17] WANG S L, CHEN Y H, WANG C L, YEN Y H, CHERN M K. Purification and characterization of a serine protease extracellularly produced by Aspergillus fumigatus in a shrimp and crab shell powder medium [J]. Enzyme and Microbial Technology, 2005, 36(5): 660-665.
[18] ESCOTT G M, HEAM V M, ADAMS D J. Inducible chitinolytic system of aspergillus fumigatus [J]. Microbiology, 1998, 144(6): 1575-1581.
[19] MARION M M. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein-dye bindiag [J]. Analytical Biochemistry, 1976, 72: 248-254.
[20] UEDA H, ARAI M. Purification and some properties of chitinases from Aeromonas sp. No.10S-24 [J]. Bioscience, Biotechnology, and Biochemistry, 1992, 56: 460-464.
(Edited by YANG You-ping)
Foundation item: Projects(50621063, 50674101) supported by the National Natural Science Foundation of China
Received date: 2008-09-15; Accepted date: 2008-11-21
Corresponding author: XIA Jin-lan, Professor, PhD; Tel: +86-731-88836944; E-mail: jlxia@mail.csu.edu.cn
- Purification and characterization of extracellular chitinase from a novel strain Aspergillus fumigatus CS-01


