
Characterization of cytochrome mutants for pellicle formation in
Shewanella onedensis MR-1
LIANG Yi-li(梁伊丽)1, 2, HE Zhi-li(贺志理)2, GAO Hai-chun(高海春)2,
QIU Guan-zhou(邱冠周)1, ZHOU Ji-zhong(周集中)1, 2, LIU Xue-duan(刘学端)1
1. School of Minerals Processing and Bioengineering, Central South University, Changsha 410083, China;
2. Institute for Environmental Genomics and Department of Botany and Microbiology, University of Oklahoma,
Norman 73019, USA
Received 9 March 2009; accepted 9 April 2009
Abstract:
Biofilm systems are effective for biosorption of metal ions. Shewanella oneidensis MR-1, a Gram-negative facultative anaerobe, is a natural pellicle-like biofilm former. The mechanisms of pellicle formation by S. oneidensis MR-1 have not yet been understood. 17 S. oneidensis MR-1 deletion mutants, including 12 c-type cytochromes were generated and tested if they were involved in pellicle formation. The results show that ?SO4666, ?SO1777, ?SO1782, ?SO2361 and ?SO2363 have varying deficiency in pellicle formation. The deletion mutant ?SO4666 cannot form a pellicle under non-shake conditions, suggesting that it may play an important role in pellicle formation by S. oneidensis MR-1. Overall, these data suggest a very complex picture of aerobic respiration by S. oneidensis MR-1.
Key words:
cytochrome; mutant; electron transport; pellicle; biofilms;
1 Introduction
Considerable studies have been focused in recent years upon the field of biosorption for the removal of metal ions from aqueous effluents[1]. Biofilms promise to be suitable systems for the treatment of metal ions since microorganisms that absorb metals stabilize the extracellular polymeric matrix by the combined action of chemical, physical and physiological phenomena that are, in some instances, linked to phenotypic variation among the constituent biofilm cells[2]. Shewanella oneidensis MR-1, a facultative Gram-negative anaerobe with a remarkable respiratory versatility, is a natural biofilm former. In recent years, this organism has attracted a great deal of interest due to its potential applications in bioremediation of metal contaminants in the environment. S. oneidensis MR-1 has been extensively studied, including the development of the canonical biofilms [3-9].
S. oneidensis MR-1 possesses a complex electron- transport system. Cytochromes, heme-containing proteins are the main components of the respiratory electron transport chains[10]. Approximately, 42 cytochrome c genes are annotated based on sequence analysis and 41 are likely to be functional in comparison with five to seven in E. coli and other enteric bacteria [11], and most of them are more or less mobile electron transfer proteins on the periplasmic side of the membrane[12]. A fraction of the Shewanella cytochrome genes have been characterized at the molecular level under anaerobic condition[13]. Since c-type cytochromes are essential for energy metabolism, their mutation will also directly affect the aerobic electron transport network. However, the role of most cytochromes under aerobic conditions remains unknown.
In this work, the mechanisms of pellicle formation in S. oneidensis MR-1 are explored. We generated and characterized 12 cytochrome mutants and 5 other mutants related to protein secretion systems and regulatory proteins, and found that some cytochromes show deficiency in pellicle formation under aerobic conditions. These data represent the first identification of genes specifically related to pellicle formation and an important step towards understanding the aerobic respiratory systems in S. oneidensis MR-1 on a genomic scale.
2 Experimental
2.1 Bacterial strains, plasmids, media, and growth conditions
S. oneidensis MR-1 and Escherichia coli strain WM3064 used for mutagenesis were grown using Luria- Bertani (LB) medium (pH 7.2) at 30 ℃ and 37 ℃, respectively.
2.2 Mutagenesis
A list of the mutants used in this study is provided in Table 1. S. oneidensis MR-1 cytochrome mutants were constructed either by fusion PCR method as described by WAN et al[14] or by a cre-lox recombination method described by MARX and LIDSTROM[15]. Other deletion mutation strains were constructed using the fusion PCR method illustrated previously[16, 17]. All deletions were verified by PCR and DNA sequencing. Each mutant could be readily distinguished by a 20 bp marker[14].
Table 1 Summary of evaluated set of cytochrome mutants, selected transport or regulator protein mutants
2.3 Complementation of ?SO4666 mutant
For complementation of ?SO4666, DNA fragment containing SO4666 and its native promoter was generated by PCR amplification with MR-1 genomic DNA as the template using primers SO4666-COM-F (AGGATCCGCAATGTCTGGTGCACTGAT) and SO4666-COM-R (AGGATCCACTTATGTTGCGGC- TGACG). This fragment was digested with BamHI and ligated to BamHI-digested pBBR1MCS-5 to form pBBR-SO4666[18], which was electroporated into WM3064. Introduction of pBBR-SO4666 into ?SO4666 was done by mating with WM3064 hosting pBBR- SO4666, and gentamycin-resistant colonies were selected. The presence of pBBR-SO4666 in ?SO4666 was confirmed by plasmid purification and restriction enzyme digestion.
2.4 Pellicle formation, measurement of growth, and quantification of pellicle
A fresh colony grown overnight on a LB plate was used to inoculate 50 mL LB and incubated in a shaker (200 r/min) to an OD600 of 0.8 at the room temperature. This culture was then diluted 500-fold with fresh LB, resulting in the starting cultures. Throughout the study, all starting cultures of S. oneidensis strains were prepared in such a way. Aliquots of 30 mL starting cultures were transferred to 50 mL Pyrex beakers equipped with side- arm stop-cocks (Lab-made). The beakers were kept still for pellicle formation at the room temperature. To separate cells in pellicle and underneath, cultures were withdrawn through the stop-cock even for collecting planktonic cells and cells from the pellicle were gathered[19]. To quantify the pellicles formed by the S. oneidensis wild-type and mutation strains, cells from pellicles were collected, suspended in 30 mL fresh LB, violently vortexed, and measured at 600 nm by a spectrometer.
2.5 Quantitative-PCR (q-PCR) estimation of biomass density during competition
Each of 12 cytochrome mutants was first grown separately to the late stationary phase in the LB medium, and then all of them were mixed together each with equal biomass, which was defined as the starting sample (C0). C0 sample was diluted by 100-fold into a flask or beaker containing 50 mL of fresh medium and grown to the late stationary phase, which was defined as a circle. Three sets of experiments differing in growth conditions, pellicle or planktonic growth under static condition and agitated culture, were performed in triplicate. The cells in pellicle and planktonic in beaker were separated by the stop-cock as we mentioned before. In agitated growth competition experiment, cytochrome mutants were co-inoculated into LB and agitated at the rate of 200 r/min for 10 cycles. The biomass of C0, circle 5 (C5) or circle 10 (C10) were collected by centrifugation and followed by DNA extraction. A certain series diluted gDNA of each 12 cytochrome mutants was used to make the standard curve by SYBR Green I fluorescence dye based q-PCR. The primers used are listed in Table 2. The PCR reactions were performed in an iQTM5 thermal cycler (BioRAD Company) and the program used was: 5 min at 94 ℃; 40 cycles at 94 ℃ for 15 s, 55 ℃ for 30 s. The biomass densities for each competitor were estimated by SYBR q-PCR using same amount of total DNA of each circle. To compare the relative fitness in each competition group, the biomass densities of C5 or C10 of each mutant were compared to that of C0. The density of certain strain was not increased or decreased if the value of log2 [(C5/C0)R5] or log2 [(C10/C0)R10] equal to 0.
Table 2 Primers used in q-RCR assays

3 Results and discussion
3.1 Pellicle formation assay
In the microbial world, existence within surface- associated structured multicellular communities is the prevailing lifestyle. Shewanella is a ubiquitous microbe suitable to live in redox interfaces in freshwater and marine, wetlands, and sediment environments where biofilm formation may provide a selective advantage. Besides canonical biofilm, Shewanella oneidensis MR-1 forms a natural pellicle-like biofilm. Two different types of cells, pellicle and planktonic co-exist in the static cultures. At the liquid-air interface, cells attach to the wall and then spread across the surface to form a layer of complex, the pellicle (Fig.1). Dissolved oxygen(DO) readings from the unshaken cultures show that DO remains stable at 0.04 mg/L. This suggests that the planktonic cells are grown under microaerobic conditions.

Fig.1 Pellicle formed by S. onedensis MR-1 in glass beaker at room temperature
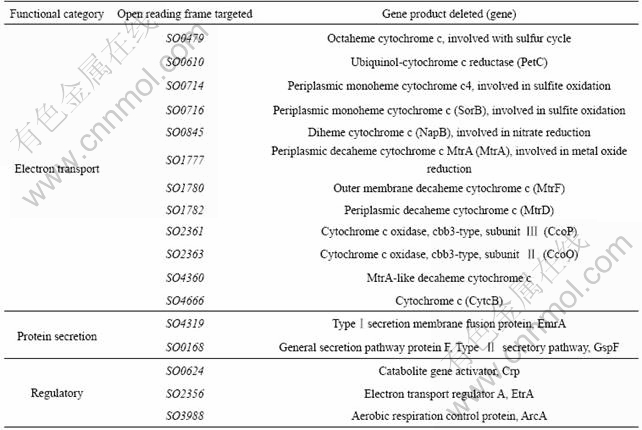
A comparison of pellicle formation at 60 h from mutants and S. onedensis MR-1 wild type is shown in Fig.2, where the level of pellicle formed by WT strain is set to 100%, and a few deletion mutants have significant effects on pellicle formation. Specifically, ?SO4666 (cytochrome c4, CytcB) showed a severe deficiency in pellicle formation. Unlike the flat, uniform biofilms formed by the WT strain, the ?SO4666 strain remained as turbid cultures composed of independent planktonic cells that were not held together by a matrix after 5-day growth. However, the complemented strain of the SO4666 deletion mutant could be fully recovered for pellicle formation. Additionally, 4 other cytochrome mutants, including ?SO1777 (periplasmic decaheme cytochrome c, MtrA), ?SO1782 (periplasmic decaheme cytochrome c, MtrD), ?SO2361 (cytochrome c oxidase, cbb3-type, subunit Ⅲ, CcoP), and ?SO2363 (cytochrome c oxidase, cbb3-type, subunit Ⅱ, CcoO) showed a partial deficiency in pellicle formation. For example, ?SO1777 formed 68.74%±4.58% of pellicle compared to the WT strain. Similarly, ?SO1782, ?SO2361, and ?SO2363 generated less than 85% of the pellicle compared to the WT strain (Fig.2). This suggests that heme-containing proteins are in close association with pellicle formation. ?SO1780 mutant strain showed no reduced ability of pellicle formation although it shared the same operon with SO1782. Mutants involved in sulfur redox reaction (?SO0479, ?SO0714, and ?SO0716), or nitrate reduction (?SO0845) had no effect on pellicle formation. Among 12 cytochrome mutants tested in this study, no mutant showed significant higher pellicle formation ability than the WT strain.

Fig.2 Relative pellicle formation by 17 (including 12 cytochrome) mutants compared to S. onedensis MR-1 wild type
Besides SO4666, other genes were found to be involved in pellicle formation. A previous study showed AggA (SO4320), type I secretion protein, increased adhesive properties of Shewanella strain[8]. Our data showed that an interruption of SO4319 (type I secretion membrane fusion protein, EmrA) resulted in severe deficiency in pellicle formation. However, another transporter mutant ?SO0168 (general secretion pathway protein F, Type II secretory pathway, GspF) sustained the ability of pellicle formation. These observations suggest that the type I secretion system is critical for pellicle formation. Consistent with previous work, the regulator mutants lacking arcA, etrA, or crp generated less than 60% of the pellicle produced by the WT[7, 20].
3.2 Static growth competition assay
In order to further understand the degree of adaptation to the environment, q-PCR was used to determine the competition ability of cytochrome mutants. In a good agreement with the independent results, the growth competition among 12 cytochrome mutants under the conditions supportive of pellicle growth revealed that ?SO4666 was undetectable at C5. At C5, the biomass of ?SO1777, ?SO1782, ?SO2361 and ?SO2363 decreased rapidly (R5<-6) compared to C0. At C10, the biomass of ?SO1777, ?SO1782, and ?SO2363 was even more dramatically decreased with the R10<-10 (Fig.3). A superior growth characteristic of SO0610 mutant (ubiquinol-cytochrome c reductase, PetC) along the course of the experiment was also observed. As it expected that slower-growing strains are overwhelmed by the faster-growing strains. The results demonstrated that SO4666 may play an important role in pellicle formation in S. oneidensis MR-1.
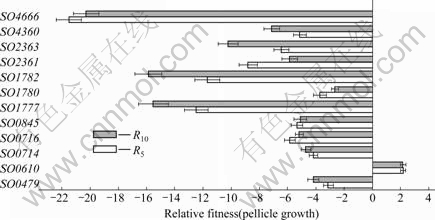
Fig.3 Pellicle growth competition among 12 cytochrome mutants
A growth competition in another set of experiment revealed that slower-growing strains in pellicle such as ?SO4666, ?SO1777, ?SO1782, ?SO2361 and ?SO2363 also grew slowly under planktonic growth condition and ?SO0610 was able to better withstand the rigorous competitive pressures (Fig.4). However, the population variations due to differences in growth rate were less apparent compared to pellicle growing conditions. At C10, the biomass of ?SO4666, ?SO1777, ?SO1782, ?SO2361 and ?SO2363 was decreased with the R10 of -6.66±0.60, -6.40±0.38, -6.37±0.38, -5.10± 0.40, and -2.47±0.35, respectively. The competition ability of ?SO4666 was similar to that of ?SO1777, ?SO1782, ?SO2361 and ?SO2363 strains under planktonic growing condition although ?SO4666 mutant was undetectable in pellicle competition set (Fig.3). It is suggested that different mechanisms may exist in pellicle formation. SO2361 and SO2363 mediate the final step of electron transfer reactions between cytochrome c and the molecular oxygen and concomitantly pumping protons across the inner membrane. SO1780 and SO1782 function as intermediate electron carriers in the Fe (Ⅲ) electron transport pathway[21]. It is possible that ?SO2361, ?SO2363, ?SO1777, ?SO1782 reduce pellicle formation by affecting growth rates under static growth conditions. ?SO4666 not only affects planktonic growth but also blocks the pellicle matrix formation at the air-liquid interface.
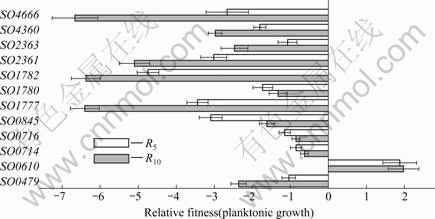
Fig.4 Planktonic growth competition among 12 cytochrome mutants
3.3 Agitated growth competition assay
S. oneidensis MR-1 has been extensively studied in anaerobic metal reduction while little is known about energy metabolism under aerobic condition. Interestingly, the results from agitated growth competition studies showed that ?SO4666 had a distinct competitive advantage over other strains, which is similar to ?SO0714 and ?SO0716 (Fig.5). Under aerobic growing conditions, ?SO1782, ?SO2361 and ?SO2363 mutants were undetectable after C10 competition and the biomass of ?SO1777, ?SO0610 and ?SO4360 decreased sharply. The results suggest that these genes may be important for aerobiosis processes, and that S. oneidensis MR-1 may have a quite complex electron transport network.

Fig.5 Agitated growth competition among 12 cytochrome mutants
Cytochrome C4 encoded by SO4666 is the least abundant of the Shewanella soluble cytochromes, and it is primarily membrane-bound like that of the Pseudomonas[22]. However, the function of SO4666 is unclear. A previous study revealed that during uranium bioremediation, a cluster of three heme-containing proteins of MR-1, MtrA-C (SO1776—SO1778) was directly localized with extracellular polymeric substance (EPS), which contained biogenic UO2 nanoparticles[23]. EPS matrix of pellicle exhibits glycocalyx-like properties and contains multiple elements of polysaccharide, DNA, and humic acid. Giving the complexity of the c-type cytochromes in MR-1, we hypothesize that the extracellular material of pellicle may be comprised, at least in part, of cytochromes or outmembrane-derived cytochromes, such as SO4666. Clearly, further studies will be required to fully understand the role of cytochromes and other factors in pellicle formation to facilitate the biosorption of metal ions.
4 Conclusions
1) The ?SO4666 mutant shows a severe deficiency in pellicle-like biofilm formation at the air-liquid interface, and is not detectable at C5 in pellicle growth competition among 12 cytochrome mutants. In addition, the biomass of ?SO4666 decreases after planktonic growth competition. The results demonstrate that SO4666 may play an important role of pellicle formation in S. oneidensis MR-1.
2) The mutants ?SO1777, ?SO1782, ?SO2361 and ?SO2363 reduce pellicle formation, and their biomass decreases after planktonic or pellicle growth competition, indicating that those genes may be involved in pellicle formation.
3) S. oneidensis MR-1 possesses a quite complex aerobic electron transport network. ?SO1782, ?SO2361 and ?SO2363 mutants are undetectable and the biomass of ?SO1777, ?SO0610 and ?SO4360 decreases sharply after agitated growth competition, which suggests that these genes may be important for aerobiosis processes. ?SO4666 has a competitive advantage over other strains in agitated growth competition.
Acknowledgements
The authors thank Shewanella Federation for supplying cytochrome mutants.
References
[1] GAVRILESCU M. Removal of heavy metals from the environment by biosorption [J]. Engineering in Life Sciences, 2004, 4(3): 219-232.
[2] HARRISON J J, CERI H, TURNER R J. Multimetal resistance and tolerance in microbial biofilms [J]. Nature Reviews Microbiology, 2007, 5(12): 928-938.
[3] BAAGGE D, HJELM M, JOHANSEN C, HUBER I, GRAMI L. Shewanella putrefaciens adhesion and biofilm formation on food processing surfaces [J]. Applied and Environmental Microbiology, 2001, 67(5): 2319-2325.
[4] de VRIENDT K, THEUNISSEN S, CARPENTIER W, de SMET L, DEVREESE B, van BEEUMEN J. Proteomics of Shewanella oneidensis MR-1 biofilm reveals differentially expressed proteins, including AggA and RibB [J]. Proteomics, 2005, 5(5): 1308-1316.
[5] THORMANN K M, DUTTLER S, SAVILLE R M, HYODO M, SHUKLA S, HAYAKAWA Y, SPORMANN A M. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP [J]. Journal of Bacteriology, 2006, 188(7): 2681-2691.
[6] THORMANN K M, SAVILLE R M, SHUKLA S, PELLETIER D A, SPORMANN A M. Initial phases of biofilm formation in Shewanella oneidensis MR-1 [J]. Journal of Bacteriology, 2004, 186(23): 8096-8104.
[7] THORMANN K M, SAVILLE R M, SHUKLA S, SPORMANN A M. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms [J]. Journal of Bacteriology, 2005, 187(3): 1014-1021.
[8] de WINDT W, GAO H C, KROMER W, van DAMME P, DICK J, MAST J, BOON N, ZHOU J Z, VERSTRAETE W. AggA is required for aggregation and increased biofilm formation of a hyper- aggregating mutant of Shewanella oneidensis MR-1 [J]. Microbiology-Sgm, 2006, 152: 721-729.
[9] TEAL T K, LIES D P, WOLD B J, NEWMAN D K. Spatiometabolic stratification of Shewanella oneidensis biofilms [J]. Applied and Environmental Microbiology, 2006, 72(11): 7324-7330.
[10] FREDRICKSON J K, ROMINE M F, BELIAEV A S, AUCHTUNG J M, DRISCOLL M E, GARDNER T S, NEALSON K H, OSTERMAN A L, PINCHUK G, REED J L, RODIONOV D A, RODRIGUES J L M, SAFFARINI D A, SERRES M H, SPORMANN A M, ZHULIN I B, TIEDJE J M. Towards environmental systems biology of Shewanella [J]. Nature Reviews Microbiology, 2008, 6(8): 592-603.
[11] BLATTNER F R, PLUNKETT G, BLOCH C A, PERNA N T, BURLAND V, RILEY M, COLLADOVIDES J, GLASNER J D, RODE C K, MAYHEW G F, GREGOR J, DAVIS N W. KIRKPATRICK H A, GOEDEN M A, ROSE D J, MAU B, SHAO Y. The complete genome sequence of Escherichia coli K-12 [J]. Science, 1997, 277(5331): 1453-1458.
[12] MEYER T E, TSAPIN A I, van DENBERGHE I, de SMET L, FRISHMAN D, NEALSON K H, CUSANOVICH M A, van BEEUMEN J J. Identification of 42 possible cytochrome c genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes [J]. OMICS-A Journal of Integrative Biology, 2004, 8(1): 57-77.
[13] BRETSCHGER O, OBRAZTSOVA A, STURM C A, CHANG I S, GORBY Y A, REED S B, CULLEY D E, REARDON C L, BARUA S, ROMINE M F, ZHOU J, BELIAEV A S, BOUHENNI R, SAFFARINI D, MANSFELD F, KIM B H, FREDRICKSON J K, NEALSON K H. Current production and metal oxide reduction by shewanella oneidensis MR-1 wild type and mutants [J]. Applied and Environmental Microbiology, 2007, 73(21): 7003-7012.
[14] WAN X F, VERBERKMOES N C, MCCUE L A, STANEK D, CONNELLY H, HAUSER L J, WU L Y, LIU X D, YAN T F, LEAPHART A, HETTICH R L, ZHOU J Z, THOMPSON D K. Transcriptomic and proteomic characterization of the fur modulon in the metal-reducing bacterium Shewanella oneidensis [J]. Journal of Bacteriology, 2004, 186(24): 8385-8400.
[15] MARX C J, LIDSTROM M E. Broad-host-range cre-lox system for antibiotic marker recycling in Gram-negative bacteria [J]. Biotechniques, 2002, 33(5): 1062-1067.
[16] GAO H C, WANG X H, YANG Z K, PALZKILL T, ZHOU J Z. Probing regulon of ArcA in Shewanella oneidensis MR-I by integrated genomic analyses [J]. Bmc Genomics, 2008, 9: 42.
[17] GAO W M, LIU Y Q, GIOMETTI C S, TOLLAKSEN S L, KHARE T, Wu L Y, KLINGEMAN D M, FIELDS M W, ZHOU J. Knock-out of SO1377 gene, which encodes the member of a conserved hypothetical bacterial protein family COG2268, results in alteration of iron metabolism, increased spontaneous mutation and hydrogen peroxide sensitivity in Shewanella oneidensis MR-1 [J]. Bmc Genomics, 2006, 7: 76.
[18] KOVACH M E, ELZER P H, HILL D S, ROBERTSON G T, FARRIS M A, ROOP R M, PETERSON K M. 4 new derivatives of the broad-host-range cloning vector pbbr1mcs, carrying different antibiotic-resistance cassettes [J]. Gene, 1995, 166(1): 175-176.
[19] BRANDA S S, VIK A, FRIEDMAN L, KOLTER R. Biofilms: the matrix revisited [J]. Trends in Microbiology, 2005, 13(1): 20-26.
[20] BUETTNER F F R, MAAS A, GERLACH G F. An Actinobacillus pleuropneumoniae arcA deletion mutant is attenuated and deficient in biofilm formation [J]. Veterinary Microbiology, 2008, 127(1/2): 106-115.
[21] PITTS K E, DOBBIN P S, REYES RAMIREZ F, THOMSON A J, RICHARDSON D J, SEWARD H E. Characterization of the Shewanella oneidensis MR-1 decaheme cytochrome MtrA [J]. Journal of Biological Chemistry, 2003, 278(30): 27758-27765.
[22] CHRISTENSEN H E. Cloning and characterization of the gene encoding cytochrome C (4) from Pseudomonas-Stutzeri [J]. Gene, 1994, 44(1): 139-140.
[23] MARSHALL M J, BELIAEV A S, DOHNALKOVA A C, KENNEDY D W, SHI L, WANG Z M, BOYANOV M, LAI I B, KEMNER K M, MCLEAN J S, REED S B, CULLEY D E, BAILEY V L, SIMONSON C J, SAFFARINI D A, ROMINE M F, ZACHARA J M, FREDRICKSON J K. Type cytochrome-dependent formation of U (IV) nanoparticles by Shewanella oneidensis [J]. Plos Biology, 2006, 324: 1333.
Foundation item: Project(50321402) supported by Chinese Science Foundation for Distinguished Group; Project(30428014) supported by National Science Fund for Distinguished Young Scholars in Hong Kong and Abroad
Corresponding author: ZHOU Ji-zhong; E-mail: jzhou@ou.edu; LIU Xue-duan; Tel: +86-731-8830546; E-mail: xueduanliu@yahoo.com
DOI: 10.1016/S1003-6326(08)60336-2
(Edited by YUAN Sai-qian)


